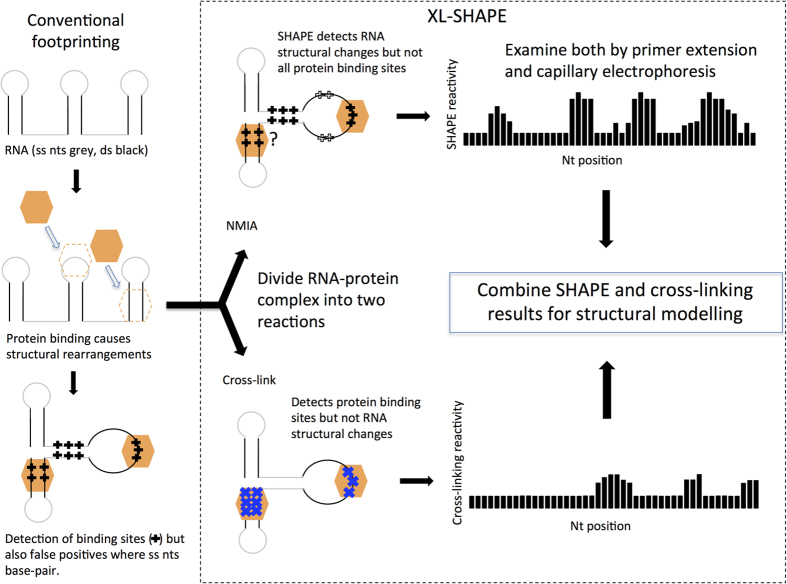
Figure 1. Footprinting vs XL-SHAPE.
Protein binding to RNA leads to structural change. Current footprinting techniques (left hand panel) can fail to differentiate between this structural change and a protein binding site-for instance in a single stranded (ss) region that becomes base-paired, where ss probing reagents are used. Such reagents can also fail to detect protein binding in double stranded (ds) regions of the RNA. In XL-SHAPE, to improve accuracy and sensitivity of RNA-protein complex modeling, samples containing the complex are divided, with one half of the sample probed with SHAPE reagents and the other half cross-linked with UV at 254 nm. SHAPE provides a detailed picture of structural changes that occur in the backbone, rather than pinpointing the interactions with proteins—these are specifically detected by cross-linking. Acylation and cross-linking sensitivity at each nt are measured using a capillary electrophoresis platform, and the data are used together to model the RNA-protein interaction.