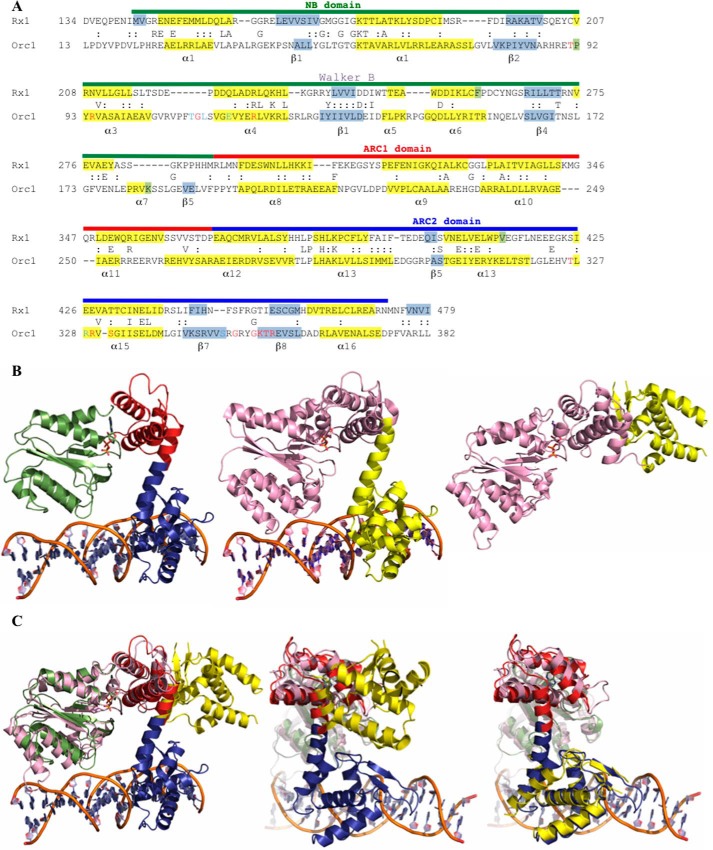
FIGURE 1.
Structural modeling of the Rx1 NB-ARC domains. A, alignment of Rx1 (residues 134–479) with Orc1 of A. pernix (PDB code 2V1U; residues 13–382). Numbers denote amino acid residue position. Sequences are in standard single-letter amino acid code, and functionally related residues between the two proteins are indicated by a colon. The Rx1 domain structure is denoted by a colored line above the Rx1 sequence and corresponds to the NB (green), ARC1 (red), and ARC2 (blue) domains. Residues in light blue contact DNA bases in the Orc1-DNA structure, whereas those in red contact DNA bases and/or the DNA backbone (45, 49). Known (Orc1) and predicted (Rx1) secondary structures (α-helix (yellow) or β-sheet (gray)) are indicated. B, structural homology model for Rx1 based on the crystal structure of DNA-bound Cdc6/Orc1 from A. pernix. Left, structural homology model of the NB-ARC domain of Rx1 (amino acids 143–4780, with associated ADP (NB domain (green), ARC1 domain (red), or ARC2 (blue)). Center, crystal structure of A. pernix Cdc6/Orc1 in complex with DNA (PDB 2V1U) (pink, amino acids 13–279; yellow, amino acids 280–399). Right, crystal structure for Cdc6/Orc1 of P. aerophilum not bound to DNA (PDB code 1FNN) (pink, amino acids 1–278; yellow, amino acids 279–388). C, comparison of the PDB 2V1U-based Rx1 homology model with the crystal structure of Cdc6/Orc1 of P. aerophilum (PDB code 1FNN). Left, complete overlay of both structures. Note that only the NB (green) and ARC1 (red) superimposes and that the ARC2 domain (blue) of Rx1 is rotated compared with the C-terminal region of Cdc6/Orc1 of P. aerophilum (yellow). Center, overlay highlighting the C-terminal ARC1 (red) and ARC2 (blue) domains of Rx1. Right, superposition of the C-terminal domain of Cdc6/Orc1 of P. aerophilum onto the Rx1 model. Domain designations are as in B.