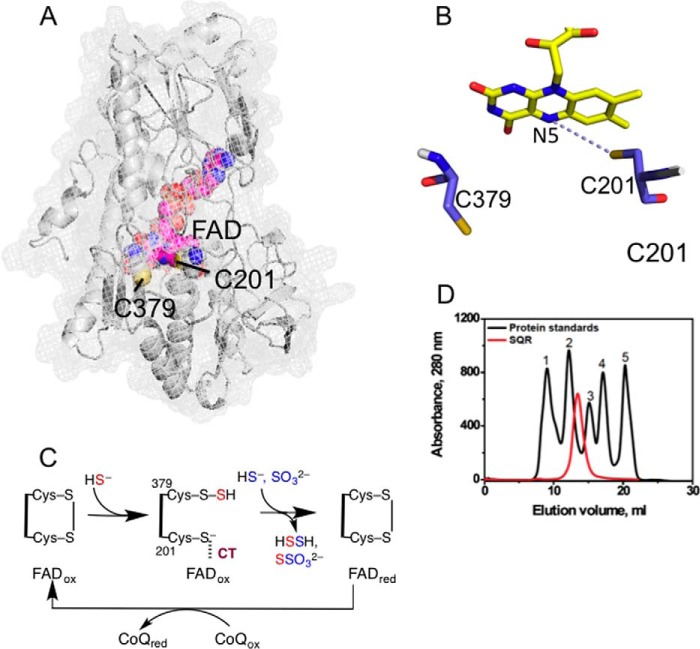
FIGURE 1.
Structure and mechanism of SQR. A, the structure of a single subunit of human SQR modeled using I-TASSER. FAD (pink) and the active site cysteine residues, Cys-201 and Cys-379, are shown in sphere representation. B, close-up of the active site showing the redox-active cofactors, flavin, and a pair of cysteine residues. C, postulated reaction mechanism of SQR in which sulfide adds into the disulfide bond in the active site, forming a persulfide intermediate and a CT complex between the liberated thiolate and oxidized FAD (FADox). The addition of sulfide or sulfite to the persulfide produces hydrodisulfide or thiosulfate and is accompanied by electron transfer to FAD and reformation of the disulfide bond. The electrons from reduced FAD (FADred) are passed to CoQ to complete the catalytic cycle. D, elution profile of SQR and gel filtration standards in 50 mm Tris buffer (pH 8) containing 0.3% DHPC, and 200 mm NaCl. The standards used were: 1, thyroglobulin (670 kDa); 2, γ-globulin (158 kDa); 3, ovalbumin (44 kDa); 4, myoglobin (17 kDa); and 5, vitamin B12 (1.35 kDa).