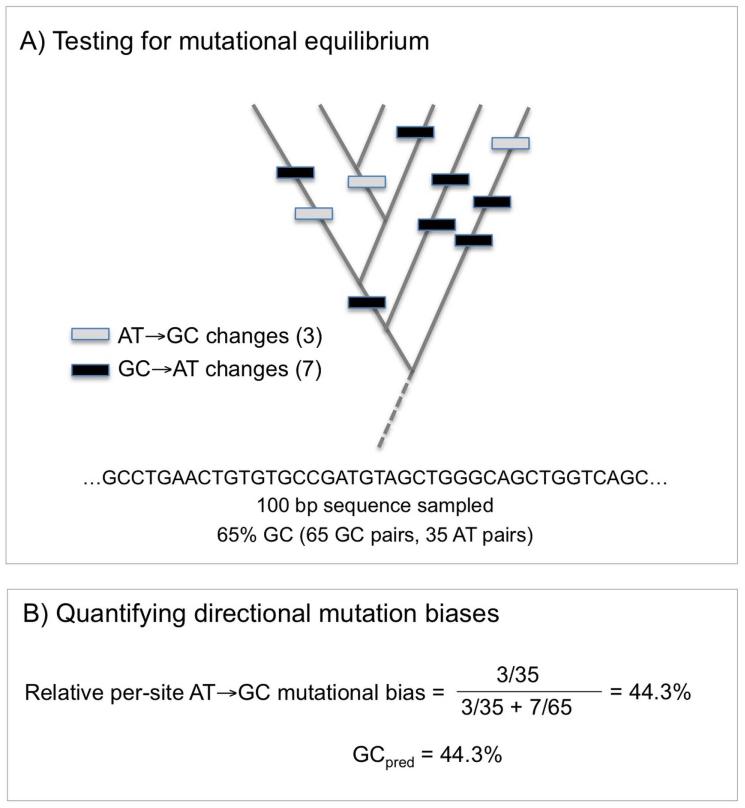
Figure 4.
Simple example of quantifying mutational spectra. Intraspecific analyses of directional base changes allow estimations of underlying mutational processes. Schematic shows a simple analysis of a 100-bp sequence. (A) Testing for mutational equilibrium addresses the question: Is current base composition explained by mutation alone? Comparing the absolute numbers of GC→AT versus AT→GC changes within species sheds light on underlying mutational pressure. If the number of AT→GC changes does not equal the number of GC→AT changes along a given sequence, then under mutation alone, the base composition of the sequence is expected to shift. By contrast, if GC→AT and AT→GC changes are equal, this implies the sequence is at mutational equilibrium. Lack of equilibrium may result from a recent change in mutational pressure or the action of natural selection favoring either GC→AT or AT→GC changes. In this example, GC→AT changes far outnumber AT→GC changes, so the sequence is not at mutational equilibrium. Rather, under mutation alone, it would become more AT-rich. (B) Quantifying directional mutation biases addresses the question: Is mutation AT or GC biased? Per-site rates of AT→GC and GC→AT changes (v and u, respectively) are obtained by dividing the absolute number of inferred changes by the frequency of the original nucleotides. In this example, v = 3/35, and u = 7/65. From these per-site rates, one can then calculate the relative AT→GC per-site mutational bias as v/(v + u). In this example, this relative per-site bias is 44.3%. Since it is less than 50%, this implies a slight GC→AT mutational bias (any given GC pair is more likely to mutate to AT, than vice versa). This value equals the base composition that would be expected under mutation alone (GCpred). In this example, the sequence is expected to be slightly AT-rich (44.3% GC) under mutation alone. The fact that the sequence is actually 65% GC suggests that selection favors AT→GC mutations.