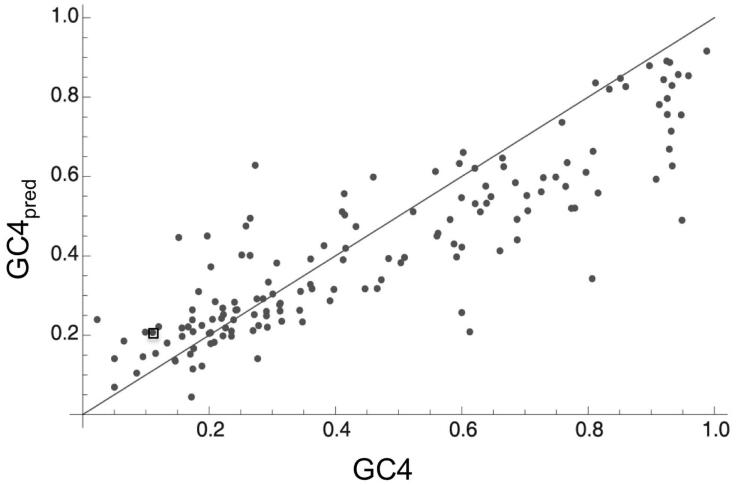
Figure 5.
The equilibrium GC content under the mutation bias model, reproduced from Hildebrand et al.133 Plot compares GC4 (current GC content at 4-fold degenerate sites) to GC4pred (GC content at 4-fold degenerate sites predicted under mutational equilibrium, estimated from intraspecific analysis), across numerous bacterial genomes spanning eight phyla. Estimates of GC4pred are based on the general approach described in Figure 4, but calculated at 4-fold generate sites only. The wide range of GC4pred across bacterial genomes suggests variation in relative per-site mutational bias. Deviation between GC4pred and GC4 indicates that mutation alone cannot explain current base composition and suggests that selection play an important role. original reference 133 for a discussion of alternative explanations to selection, such as biased gene conversion (BGC), and evidence against these alternatives. While this figure suggests that AT-rich bacteria also show a deviation between GC4pred and GC4, and in the opposite direction (less GC-rich than expected under mutation), this deviation was not significant among AT-rich genomes after adjusting for the infinite sites model.133 The point corresponding to the aphid mutualist Buchnera is enclosed in a square, for reference. Other AT-rich bacteria shown include intracellular pathogens.