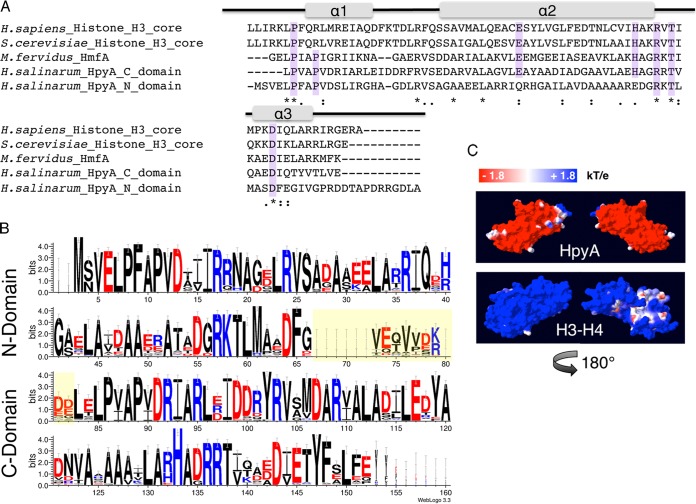
FIG 1 .
Haloarchaeal species encode a single conserved histone heterodimer. (A) Alignment of histone protein sequences: the eukaryotic H3 core domains from Homo sapiens and Saccharomyces cerevisiae, HMfA from Methanothermus fervidus, and each of two histone fold domains of H. salinarum HpyA. Residues essential for histone function or cell viability in the Eukarya and/or Archaea are highlighted in purple. Gray bars indicate the locations of the three alpha-helical regions that comprise the histone fold domain of the canonical eukaryotic H3. Asterisks, fully conserved residues; colons, strongly similar residues; periods, weakly similar residues. (B) Consensus logo representing histone sequence conservation between haloarchaeal species. The linker region between histone fold domains (yellow-shaded residues) is variable in length and sequence. Residue colors are as follows: red, acidic; black, uncharged; blue, basic. (C) Space-filling representation of the surface charge of H. salinarum HpyA and in silico fusion of the S. cerevisiae H3 to H4 dimer modeled onto the M. kandleri histone HMk crystal structure (10). Blue indicates a positive charge, whereas red indicates a negative charge (see scale at top).