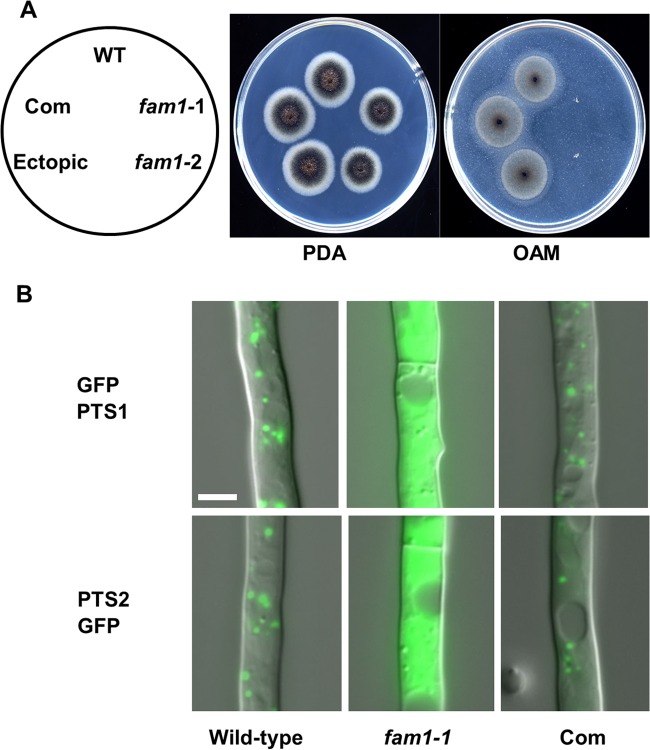
FIG 2 .
C. orbiculare fam1 disruption mutants are deficient in peroxisome biogenesis. (A) The fam1 mutants are defective in fatty acid utilization. Colonies were grown for 5 days on PDA or 12 days on oleic acid-containing medium (OAM). The strains tested were the wild-type (WT) strain, fam1 mutants (fam1-1 and fam1-2), fam1-1 complemented strain (Com), and ectopic transformant. The locations of strains on the plates are indicated in the left-hand panel. (B) Mislocalization of GFP-PTS1 and PTS2-GFP proteins in the fam1 mutant. Vegetative hyphae of the wild-type, fam1-1 mutant, and the fam1-1 complemented strain (Com) expressing GFP-PTS1 or PTS2-GFP were grown for 24 h on glass slides in liquid medium and observed by differential interference contrast (DIC) and epifluorescence microscopy. Photos are merged images. Bar, 5 µm.