Fig. 1.
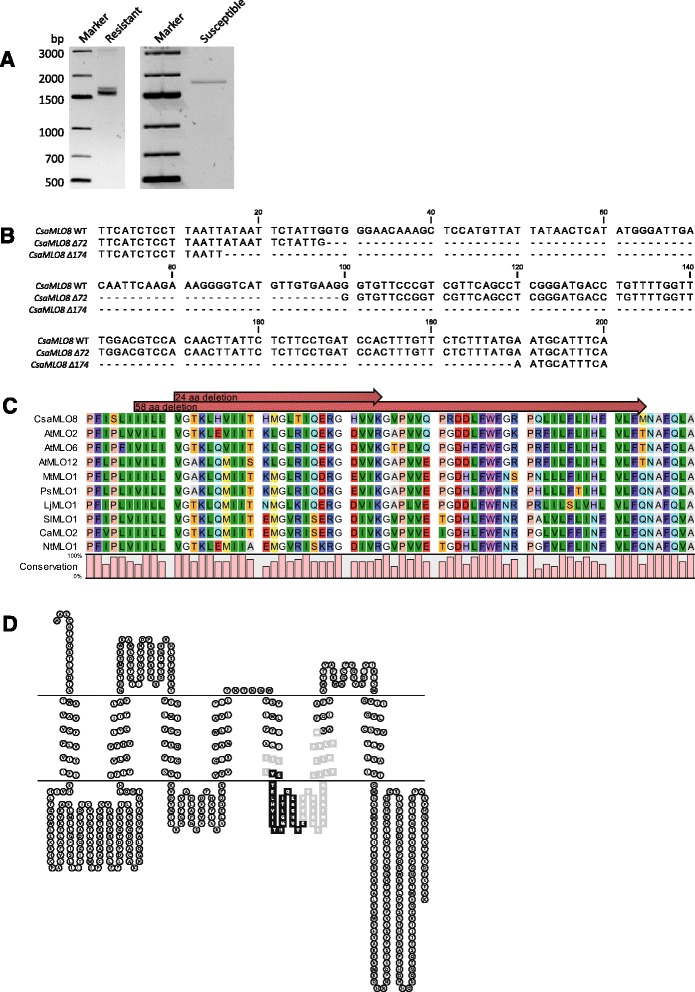
Characterization of CsaMLO8 alleles from resistant and susceptible cucumber genotypes. a cDNA of resistant (left panel) and susceptible (right panel) cucumber genotypes was used as template for PCR with CsaMLO8 specific primers. Amplified products were analysed on 1.25 % agarose gels. Whereas the product amplified from cDNA of the susceptible genotype gives a single band of the expected size, cDNA of the resistant genotype results in two separate bands, both of a smaller size than expected. b Full length CsaMLO8 amplified from cDNA from susceptible and resistant cucumber genotypes was sequenced. A partial alignment is shown between the (wild-type) sequence as obtained from the susceptible genotype and the sequences from two deletion variants (∆72 and ∆174) obtained from the resistant genotype. Numbers are relative to the start of the alignment. c Partial alignment of the CsaMLO8 protein and other proteins encoded by clade V MLO S-genes of several species. Amino acid residues are coloured according to the RasMol colour scheme. The 24 and 58 amino acid residues deleted in the proteins encoded by the ∆72 and the ∆174 variants of CsaMLO8 are indicated by red arrows. A bar graph underneath the alignment indicates the conservedness of each amino acid position. d Graphic representation of the transmembrane structure of the predicted CsaMLO8 protein, determined using HMMTOP 2.1 [30]. The plasma membrane is indicated by two horizontal lines. Amino acid residues highlighted in black are predicted to be deleted in the protein encoded by the ∆72 variant of the CsaMLO8 gene, residues highlighted in black and grey are predicted to be deleted in the protein encoded by the ∆174 variant of the CsaMLO8 gene
