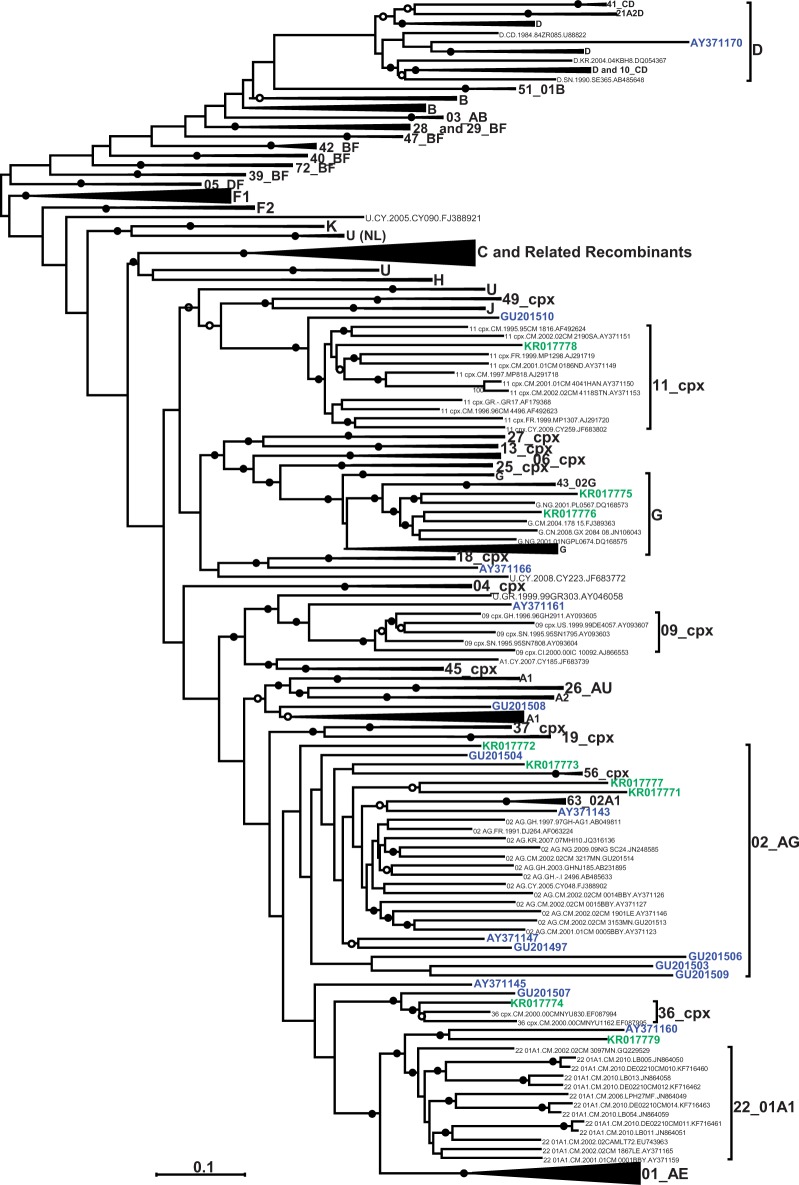
Figure 1.
Maximum likelihood tree indicating the phylogenetic placement of the new near full-length genome sequences characterised in this study (in green) plus 15 other previously identified divergent HIV-1M genomes from Cameroon (in blue) and a representative selection of near full-length sequences from all published subtypes, CRFs and unclassified sequences available in the LANL database (http://hiv-web.lanl.gov/content/hiv-db) in June 2014. Some clades have been collapsed for the sake of clarity. A larger tree with un-collapsed clades is available on request. The tree was constructed with 500 full ML bootstrap replicates using RAxML. Solid and open circles indicate branches with >70 and 50% bootstrap support, respectively. The tree was midpoint rooted