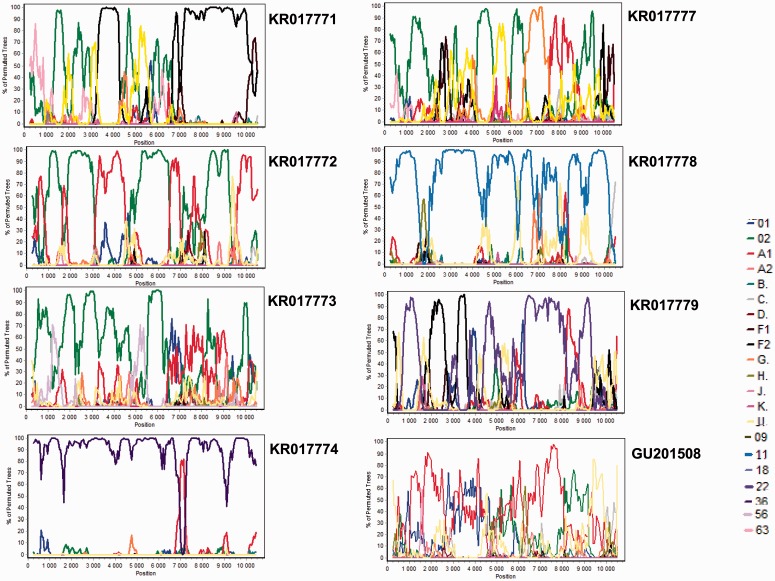
Figure 2.
Recombination analysis of the newly sequenced strains plus a previously described strain GU201508. The multiple genome alignment used to calculate the consensus reference sequences (90% threshold) included the same references as used for the phylogenetic tree analysis. The newly sequenced and previously described viruses were queried against strains from subtypes A to D, F to H, J, K, CRF01_AE, and CRF02_AG and, in some cases, lineages of viruses they were most closely related to in the ML tree; the reliability of plot topologies was assessed by bootstrapping with 500 replicates, and a sliding window of 500 bp advancing with 50-bp increments