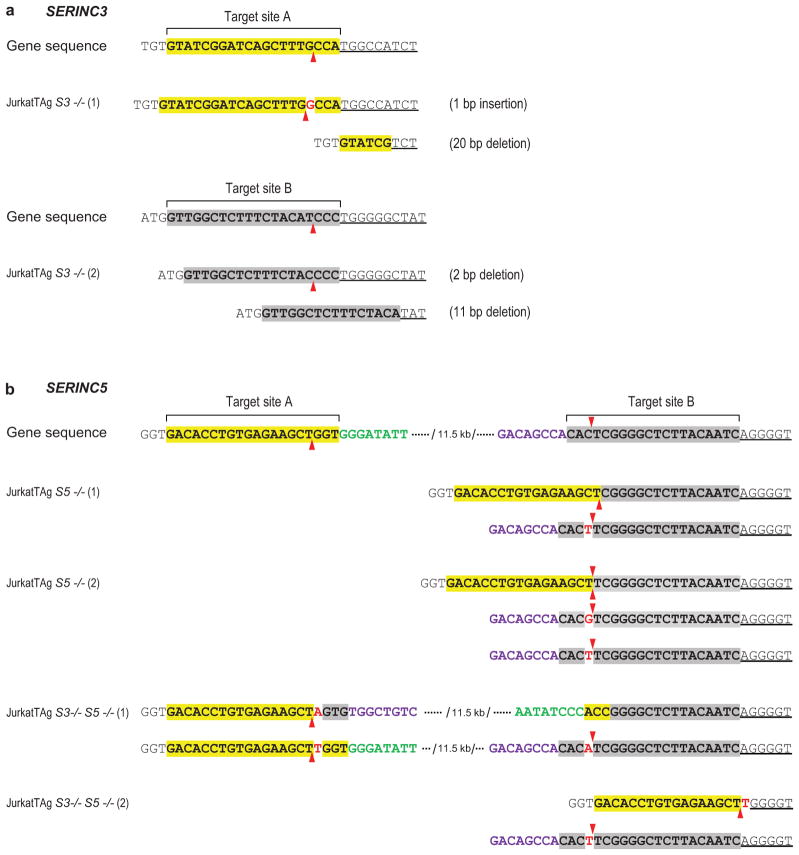
Extended Data Figure 8. Characterization of JurkatTAg KO cells.
a, Mutant SERINC3 alleles identified in SERINC3 KO clones. b, Mutant SERINC5 alleles identified in SERINC5 KO and SERINC3/5 double-KO clones. The sgRNA target sites are highlighted, and the predicted Cas9 target sites are indicated by arrowheads. Inserted nucleotides are in red. One of the two mutated SERINC5 alleles in JurkatTAg S3−/− S5−/− (1) cells has an inversion between sgRNA target sites A and B. JurkatTAg S5−/− (2) cells harbor 3 mutated SERINC5 alleles. All mutations cause frameshifts and/or large deletions of coding sequence. No WT alleles were detected in any of the KO clones.