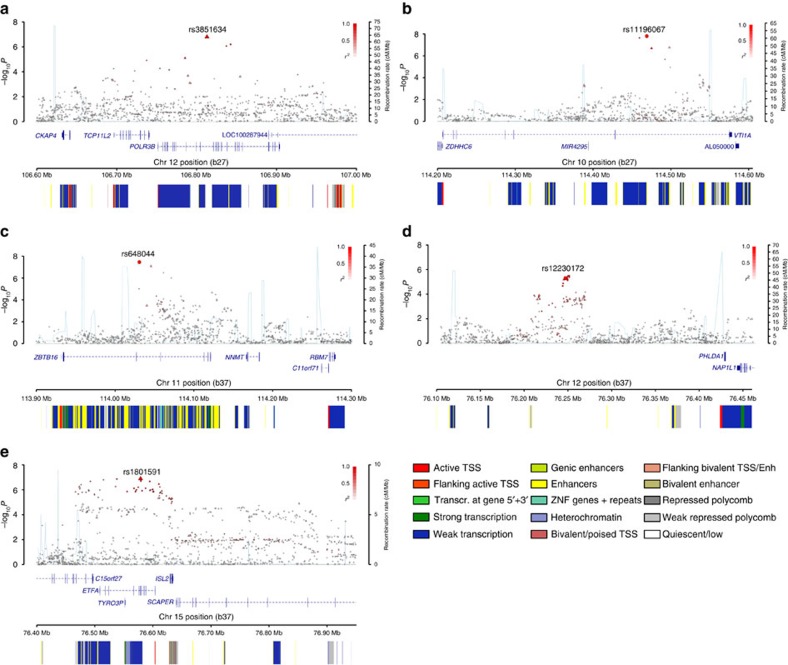
Figure 2. Regional plots of discovery-phase association results, recombination rates and chromatin state segmentation tracks for five glioma-risk loci.
Results for: (a) 12q23.33, rs3851634 (GBM); (b) 10q25.2, rs11196067 (non-GBM); (c) 11q23.2, rs648044 (non-GBM); (d) 12q21.2 rs12230172 (non-GBM); and (e) 15q24.2, rs1801591 (non-GBM). Plots show discovery association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates. The −log10 P values (y axes) of the SNPs are shown according to their chromosomal positions (x axes). The lead SNP in each combined analysis is shown as a large circle or triangle (if imputed or directly genotyped, respectively) and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top genotyped SNP, white (r2=0) to dark red (r2=1.0). Genetic recombination rates, estimated using HapMap samples from Utah residents of western and northern European ancestry (CEU), are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of University of Carolina, Santa Cruz (UCSC) genes and transcripts mapping to the region of association. Genes have been redrawn to show their relative positions; therefore, maps are not to physical scale. Below each plot is a diagram of the exons and introns of the genes of interest, the associated SNPs and the chromatin state segmentation track (ChromHMM) for H1 neural progenitor cells derived from the epigenome roadmap project, as per legend. TSS, transcriptional start sites.