Abstract
Intrinsically disordered proteins (IDPs) perform their physiological role without possessing a well-defined three-dimensional structure. Still, residual structure and conformational dynamics of IDPs are crucial for the mechanisms underlying their functions. For example, regions of transient secondary structure are often involved in molecular recognition, with the structure being stabilized (or not) upon binding. Long-range interactions, on the other hand, determine the hydrodynamic radius of the IDP, and thus the distance over which the protein can catch binding partners via so-called fly-casting mechanisms. The modulation of long-range interactions also presents a convenient way of fine-tuning the protein’s interaction network, by making binding sites more or less accessible. Here we studied, mainly by nuclear magnetic resonance spectroscopy, residual secondary structure and long-range interactions in nonstructural protein 5A (NS5A) from hepatitis C virus (HCV), a typical viral IDP with multiple functions during the viral life cycle. NS5A comprises an N-terminal folded domain, followed by a large (∼250-residue) disordered C-terminal part. Comparing nuclear magnetic resonance spectra of full-length NS5A with those of a protein construct composed of only the C-terminal residues 191–447 (NS5A-D2D3) allowed us to conclude that there is no significant interaction between the globular and disordered parts of NS5A. NS5A-D2D3, despite its overall high flexibility, shows a large extent of local residual (α-helical and β-turn) structure, as well as a network of electrostatic long-range interactions. Furthermore, we could demonstrate that these long-range interactions become modulated upon binding to the host protein Bin1, as well as after NS5A phosphorylation by CK2. As the charged peptide regions involved in these interactions are well conserved among the different HCV genotypes, these transient long-range interactions may be important for some of the functions of NS5A over the course of the HCV life cycle.
Introduction
Intrinsic conformational disorder is a prevalent feature among viral proteins because flexibility confers a functional advantage for binding to multiple partners (1–4). Viral proteins, containing a significant extent of disordered regions, typically interact via transient, low-affinity interactions that are often regulated by posttranslational modifications, such as phosphorylation (5). Posttranslational modification sites are preferably located in disordered protein regions in order to be easily accessible to the required enzymes (6–9). Another advantage conferred by intrinsic disorder to regulatory proteins, especially if they are membrane-anchored, is the larger hydrodynamic radius that allows easier establishment of contacts with the binding partners by the so-called fly-casting mechanism (10).
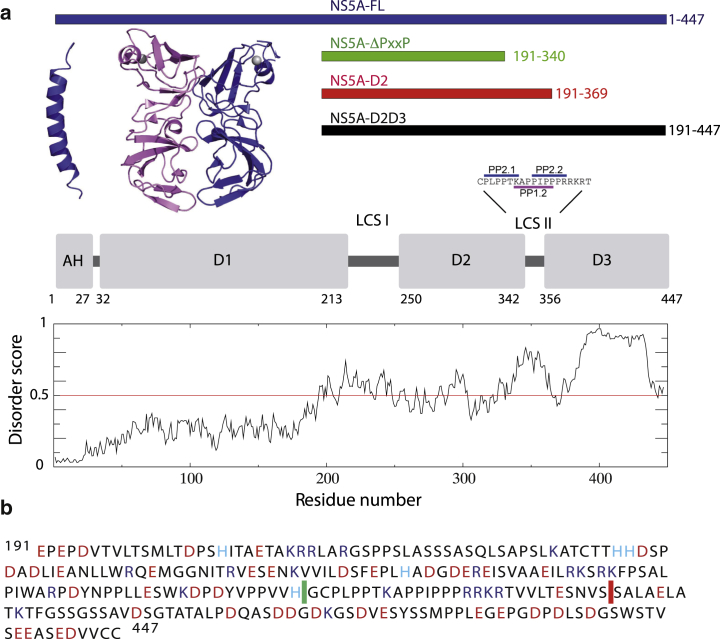
Hepatitis C virus (HCV) has a small genome encoding only 10 different viral proteins. Interestingly, most of these proteins have regions with increased flexibility or are intrinsically disordered (11). One of them, nonstructural protein 5A (NS5A), is a typical multifunctional, intrinsically disordered protein (IDP) that plays a dual role for the viral life cycle: it is an essential part of the viral replication machinery, and it is also indispensable for viral particle assembly taking place at lipid droplets. NS5A interacts with a variety of other viral and host proteins in order to recruit host proteins required for viral replication and/or particle assembly, and to perturb the host cellular machineries. An example is its interaction with the tumor suppressor protein Bin1 (amphiphysin 2) (12). NS5A contains a proline-rich region made up of three polyproline motifs, referred to as a low-complexity sequence (LCS) II (Fig. 1 a), which has been identified as a binding site for SH3 domains of various host proteins (13). Deletion of this proline-rich region results in HCV phenotypes that are incapable of replication.
Figure 1.
(a) Domain organization of NS5A from HCV genotype 1b, and structural disorder score as predicted by IUPRED (14). The structures of the amphipathic N-terminal helix (15) and the domain D1 (16) are plotted on top together with the definition of the four NS5A constructs (indicated by bars) used for this study. LCS denotes low complexity sequences located between the domains. The sequence of LCS II is expanded and the three polyproline motifs located in this region are shown. (b) Amino-acid sequence (one-letter code) of the intrinsically disordered region of NS5A (191–447) corresponding to the NS5A-D2D3 construct. Negatively charged residues (red); positively charged residues (blue). The C-termini of the shorter constructs are indicated for NS5A-ΔPxxP (green line) and NS5A-D2 (red line). To see this figure in color, go online.
NS5A is anchored to host endoplasmic reticulum membrane alterations by a short N-terminal amphipathic helix of ∼30 residues in length (17) that is followed by three domains located in the cytosol (18). The structure of the first domain (D1) has been elucidated by x-ray crystallography. A growing number of crystal structures of domain 1 from genotypes 1a and 1b, with four different dimeric forms, are available as of this writing (16,19,20). These high-resolution structures revealed a potential RNA binding groove (21). Together with biochemical data on RNA binding to NS5A (22), this led to the hypothesis that one function of NS5A is the translocation of the viral RNA from the site of replication to the site of particle assembly (23). The remainder of the NS5A protein, made up of domains 2 (D2) and 3 (D3), is intrinsically disordered as demonstrated both experimentally (24,25) and by computational analysis of the intrinsic disorder in the HCV genome (11). In addition, it has been shown that D2 is required for RNA replication (26) while domain 3 is indispensable for particle assembly (27). Recent high-throughput pharmacological inhibitor screening recognized NS5A as a promising therapeutic target for the treatment of HCV (28). However, up to now, little has been known about the exact molecular mechanisms of its action—and detailed structural studies have yet to be performed to reveal them. A 2012 study does provide some molecular details on the interaction of NS5A with NS5B, the viral RNA polymerase, and the host factor cyclophilin A (CypA), both of which are crucial for viral RNA replication. The NS5A binding sites for both NS5B and CypA were localized in D2 (29). However, no structural details about the complexes are available as of this writing, and no information whether the binding requires preformed molecular recognition elements in this highly disordered D2 domain.
NS5A is a phosphoprotein, existing in the cell as two major, differently phosphorylated forms, termed basal- and hyper phosphorylated. The main phosphorylation sites have been localized in the intrinsically disordered region of the protein (30). So far, protein kinases CK2, CK1α, PKA, and Plk1 have been shown—using kinase inhibitors and gene silencing—to be physiologically relevant for NS5A phosphorylation in vivo (21,27,31,32). To date, only a very few of the phosphorylation sites have been localized, and comprehensive data about eventual genotypic differences is lacking. There is ample evidence in the scientific literature for the importance of NS5A phosphorylation for HCV replication and pathogenesis. For example, the inhibition of certain kinases has been shown to impact viral replication (33), while the HCV drug daclatasvir acts on the HCV life cycle by altering the phosphorylation state of NS5A (34). Furthermore, it is known that phosphorylation by protein kinase CK2 of serine residues in D3 is required for binding to the HCV core protein, and for particle assembly (27,35,36). A molecular explanation of how NS5A phosphorylation affects NS5A structure and function is still missing, as well as detailed knowledge on the kinases involved and the phosphorylation sites.
Up to now, nuclear-magnetic resonance (NMR) studies of NS5A from different genotypes have been focusing on small fragments of the disordered part of the protein, typically made up of either the D2 or D3 domain. While such protein constructs facilitate sample handling, as well as NMR data recording and analysis, the question remains whether such truncated protein constructs correctly reproduce the conformational and functional properties of the protein. In particular, little is known about the synergistic action among the three domains in the context of membrane-anchored full-length NS5A, and how the presence of long-range interdomain interactions influences the conformational properties of the protein in solution. Here, we report NMR data for the entire disordered region of NS5A (residues 191–447) from genotype 1b, both in isolation as well as in the context of the full-length protein. This data has allowed us to characterize transient structural features in this functionally important part of the NS5A protein. In particular, we identified electrostatically driven long-range interactions that are modulated by phosphorylation, as well as by binding to the SH3 domain of the tumor suppressor protein Bin1.
Materials and Methods
Recombinant protein production and purification
The NS5A-D2 and NS5A-ΔPxxP constructs were produced and purified as described in Feuerstein et al. (37) and Schwarten et al. (38). The gene encoding for NS5A-D2D3 was purchased from GENEART (Regensburg, Germany), and subcloned into a modified pET28 vector, where the original thrombin cleavage site was exchanged against a Tobacco Etch Virus (TEV) cleavage site. This results in a TEV protease cleavable His-tag at the N-terminus. Escherichia coli BL21 DE3 cells were transformed with this vector and grown at 37°C in M9 medium with isotopically enriched 15N NH4Cl (1 g/L) and 13C-glucose (2 g/L) until reaching an OD600 between 0.6 and 0.8, respectively. At this point, expression was induced by addition of 1 mM IPTG (isopropyl β-D-thiogalactopyranoside) and the cells were incubated at 25°C for additional 4 h. The cells were harvested by centrifugation and subsequently lysed by sonication in the buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl, 20 mM imidazole, pH 8.0). The supernatant of the cell lysis was passed through a Ni-NTA column under denaturing conditions. The protein was eluted with increasing concentrations of imidazole. After dialysis to cleavage buffer (50 mM K2HPO4/KH2PO4, 200 mM NaCl, 2 mM 2-mercaptoethanol, pH 7.0) the His-tag was cleaved by TEV protease for 2 h at room temperature. The resulting protein was separated from the uncleaved protein and the protease by a second Ni-NTA chromatography step. A gel filtration step (50 mM K2HPO4/KH2PO4, 200 mM NaCl, 2 mM 2-mercaptoethanol, pH 8.0) yielded the pure protein. As a final step, the protein was dialyzed in the NMR buffer (50 mM K2HPO4/KH2PO4, 20 mM NaCl, 2 mM TCEP, pH 6.5).
NS5A-FL was produced as follows: a synthetic gene coding for HCV NS5A-FL (UniProt:O92972) genotype 1b was cloned into the vector pET41c with His-tag at the C-terminus. BL21DE3 star cells were transformed with the plasmid DNA and grown at 37°C in M9 medium supplemented with 13C glucose and 15N ammonium chloride. Expression was induced at an OD600 of 0.6–0.8 by adding 1 mM IPTG and the cells were further grown overnight at 20°C. Cells were resuspended in lysis buffer (40 mM Tris pH 8.0, 100 mM NaCl, 10 mM 2-mercaptoethanol, 5% glycerol, and protease inhibitor) and sonicated on ice. The pellet was recovered from centrifugation at 15,000 g and dissolved in 3% sarkosyl. Then the solution was subjected to 40,000 g and the supernatant was dropwise flash-diluted into the refolding buffer (40 mM Tris pH 8.0, 100 mM NaCl, 2 mM 2-mercaptoethanol, and 0.05% Fos-Choline-12). The supernatant was passed through Ni-NTA. The eluted fraction with imidazole was further injected into a gel filtration column and washed with buffer (20 mM MES, 100 mM NaCl, 5 mM 2-mercaptoethanol, 0.1% Fos-Choline-12, pH 6.5). NS5A-FL/NAPol was prepared as reported in Bazzacco et al. (39). NAPols are totally nonionic, amphiphilic polymers synthesized by free-radical polymerization of acrylamide monomers in the presence of a thiol-based transfer agent (40). Homopolymeric NAPols result from polymerization of an amphiphilic monomer carrying two glucose moieties and a single undecyl alkyl chain (41). For the NAPol used in this study, NA13 (batch MB155), the average molecular mass is ∼13 kDa. Briefly, after incubation with NAPol for 20 min at room temperature, n-dodecylphosphocholine was removed by incubation with Bio-Beads SM-2 nonpolar polystyrene adsorbents (Bio-Rad, Hercules, CA) for 2 h at room temperature. Three cycles of dilution and concentration were performed in order to remove any traces of n-dodecylphosphocholine.
NMR spectroscopy
The NMR sample concentration was 120 μM, except for NS5A-FL, where maximal sample concentration was limited to 90 μM. Typically, 5-mm tubes (Shigemi, Allison Park, PA) were used containing 300 μL of NS5A sample. All experiments were performed on high-field NMR spectrometers equipped with cryogenically cooled triple-resonance probes (HCN) and pulsed z-field gradients, using either a model No. VNMRS 800 (Agilent Technologies, Santa Clara, CA) or an Avance III HD 700-, 850-, and 950-MHz instruments (Bruker, Billerica, MA). For titration and phosphorylation real-time kinetics experiments, two-dimensional 1H-15N BEST-TROSY (42) spectra were recorded. Assignment of NS5A-D2D3 at 25°C was achieved from a series of BT-HNcoCACB, BT-iHNCACB, BT-hNcocaNH, and BT-HNcocaHA experiments (43). NMR spectra were processed using the software NMRPIPE (National Institutes of Health, Bethesda, MD; http://spin.niddk.nih.gov/NMRPipe/) (44), and analyzed using the software CCPNMR (http://www.ccpn.ac.uk/software/analysis) (45). Random coil chemical shifts were calculated using the Poulsen IDP/IUP random-coil chemical shifts web server (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health; http://spin.niddk.nih.gov/bax/nmrserver/Poulsen_rc_CS/) and β-turn probabilities were calculated using the MICS server, located at http://spin.niddk.nih.gov/bax.
15N T1, T2, and heteronuclear NOE (HetNOE) data were measured at two magnetic field strengths (16.5 and 18.8 T) and two sample temperatures (5 and 25°C). 15N relaxation experiments were performed with standard Bruker (at 700 MHz) and Agilent Technologies (at 800 MHz) library sequences. Reduced power spectral density mapping was performed according to Farrow et al. (46) using a PYTHON script (www.python.org). Uncertainties in the calculated spectral density values were estimated from Monte Carlo simulations, using the errors from the exponential fit to the relaxation data for T1 and T2 and the error estimated from the spectral noise for HetNOE.
Titration experiments of NS5A-D2D3 with Bin1-SH3 were performed starting from a 120 μM NS5A-D2D3 sample, and a 1.2 mM Bin1-SH3 stock solution, both dialyzed against the same NMR buffer. Bin1-SH3 was then added gradually to produce 1:0.5 and 1:1 [NS5A]:[SH3] complexes, and chemical shift changes were monitored by recording two-dimensional 1H-15N BEST-TROSY spectra. Finally, BT-HNCA and BT-HNCO spectra as well as 15N relaxation data (T1, T2, HetNOE) were recorded on the final 1:1 complex.
Measurement of phosphorylation kinetics was performed on NS5A-D2D3 samples dialyzed into the phosphorylation buffer (100 mM Tris, 10 mM MgCl2, 50 mM KCl, 5 mM DTT, pH 7.5). ATP was added just before the start of the reaction to a final concentration of 5 mM, and 4000 U CK2 kinase (New England BioLabs, Ipswich, MA) was added to the 300 μL sample. This time point was noted as the start of the phosphorylation reaction. The sample was transferred into a tube (Shigemi) and inserted into an 850 MHz NMR spectrometer (Bruker). A series of two-dimensional 1H-15N BEST-TROSY spectra were recorded during 12 (or 6.3) h, using an interscan delay of 100 ms, and an experimental time of 10 min. Peak intensities were then fitted as a function of phosphorylation time by a PYTHON script using the CURVEFIT function of the SCIPY module (www.scipy.org).
Small-angle x-ray scattering
Small-angle x-ray scattering (SAXS) measurements were performed on the NS5A-D2D3, NS5A-ΔPxxP, and NS5A-D2 constructs dissolved in the NMR buffer at the DESY X33 beamline at the European Molecular Biology Laboratory (EMBL, Hamburg, Germany), as described in Feuerstein et al. (37) for the NS5A-D2 construct. Briefly, SAXS data were recorded for different protein concentrations (90, 50, and 30 μM) at 5°C. The background scattering of the buffer, measured separately, was subtracted from the scattering of the protein sample. Unfortunately, the SAXS data obtained for NS5A-D2D3 were contaminated by scattering contributions from protein aggregates. Therefore, these data were not used for further analysis. For data analysis, the program EOM (Ver. 2.0) (47) was used from the ATSAS package (EMBL; http://www.embl-hamburg.de/biosaxs/software.html), which avoids overfitting by minimizing the number of selected conformers that are sufficient to reproduce the scattering data.
Results
To characterize the conformational properties of the intrinsically disordered C-terminal part of NS5A by NMR spectroscopy, we have designed four NS5A constructs of different length, as depicted in Fig. 1 a: 1) a full-length NS5A (NS5A-FL) construct; 2) a 256-residue NS5A fragment made up of residues 191–447 (NS5A-D2D3) corresponding to the entire disordered region as predicted by the software IUPRED (http://iupred.enzim.hu/) (14); 3) a shorter construct from residue 191 to 369 (NS5A-D2) that lacks the D3 domain; and 4) NS5A fragment 191–340 (NS5A-ΔPxxP) that misses D3 as well as the polyproline region in LCS II. The amino-acid type composition of the disordered C-terminal part of NS5A is shown in Fig. 1 b. NMR assignments and structural characterization of the latter two shorter constructs have already been reported elsewhere (37,38), and these data are used here only for the purpose of comparison with the longer NS5A constructs.
NMR investigation of NS5A-FL
A major complication for the study of full-length NS5A (NS5A-FL) by solution NMR techniques is that NS5A-FL is not soluble in pure aqueous buffers. First attempts using liposomes or nanodiscs (48) as solubilizing agents failed, as NS5A-FL precipitated heavily in both cases. Therefore we used detergent micelles to solubilize NS5A-FL. A range of detergents were tested, and all of them yielded NMR spectra of moderate to good quality, mainly differing in the number of detected signals in a 1H-15N correlation spectrum, as well as their line width and intensity. The differential NMR line broadening observed for NS5A-FL dissolved in various detergents points toward specific interactions between residues in NS5A-D2D3 and the detergent. To evaluate the effect of different detergents, we have recorded 1H-15N spectra of NS5A-D2D3 after adding identical amounts of detergent as used for solubilization of NS5A-FL (Fig. S1 in the Supporting Material). High-quality NMR spectra were obtained for nonionic detergents with glycosides (i.e., DDM, OG), as well as in nonionic amphipols called NAPols (40,41), while extensive line broadening for a large number of NS5A-D2D3 residues was observed for zwitterionic detergents (i.e., Fos-Choline-12, lauryldimethylamine oxide) that are often used to dissolve well-folded membrane proteins. The best results in terms of protein solubility, stability, and minimal line broadening was obtained using NAPols as medium for NS5A-FL.
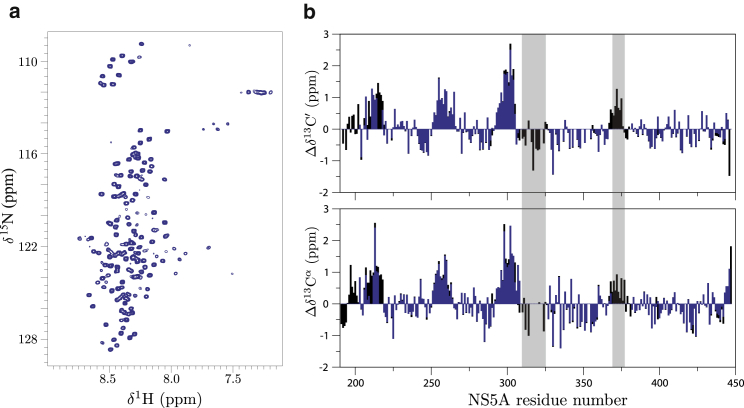
Comparison of the NS5A-FL spectrum with the corresponding spectrum recorded on NS5A-D2D3 shows that the observed correlation peaks originate, as expected, from the highly disordered C-terminal part of the protein (Fig. 2 a). The structured D1 and the N-terminal α-helix are not observed due to their slow rotational tumbling in solution. NMR assignment of NS5A-FL dissolved in NAPol was obtained mainly by analogy with the available assignment for the NS5A-D2D3 construct, and independently confirmed and completed using a series of three-dimensional BEST-TROSY HNC experiments (43). NMR signals are detected for the whole disordered C-terminal region starting with residue T204, indicating that this part of the protein also displays a high degree of conformational flexibility in the context of full-length NS5A. Chemical shift differences between NS5A-FL and NS5A-D2D3 are minor and localized to two regions (301–321 and 363–382) that also show shifts when adding NAPol to NS5A-D2D3 (Fig. S2). Therefore these chemical shift changes can be safely attributed to the interaction of NAPol with specific residues in NS5A-D2D3. No indication for a specific interaction between NS5A-D2D3 with either D1 or the N-terminal helix was observed. As a first consequence of these observations, we can conclude that the NS5A-D2D3 construct presents a valid model system for investigating the conformational dynamics in the intrinsically disordered part of full-length NS5A, as well as the interaction with binding partners and the effect of phosphorylation on the conformational properties of the protein. Furthermore, we have established a valid experimental model of membrane-anchored full-length NS5A that is amenable to solution-state NMR investigation of the highly disordered C-terminal half of NS5A, and that can be used to validate results obtained on the shorter NS5A-D2D3 construct whenever needed. Of course, care has to be taken when analyzing NS5A-FL data concerning the two protein regions that have been shown to interact with the NAPol.
Figure 2.
NMR data recorded on NS5A-FL. (a) 1H-15N BEST-TROSY spectrum recorded at 5°C and 950 MHz. (b) Overlay of 13C′ and 13Cα secondary chemical shifts for NS5A-D2D3 (black) and NS5A-FL (blue). Peptide regions showing missing or low cross-peak intensities in NS5A-FL are highlighted (gray bars). To see this figure in color, go online.
Structural characterization of the intrinsically disordered NS5A-D2D3 fragment
1HN, 1Hα, 13Cα, 13Cβ, 13CO, and 15N resonance assignment of NS5A-D2D3 at two temperatures, 25 and 5°C, was obtained from a series of three-dimensional BEST-TROSY HNN and HNC correlation experiments (43), as well as HADAMAC (49,50) spectra, similar to our previous work on NS5A-D2 (37,51). Complete backbone NMR assignment was obtained, except for proline residues, and deposited with the BMRB (accession No. 26549).
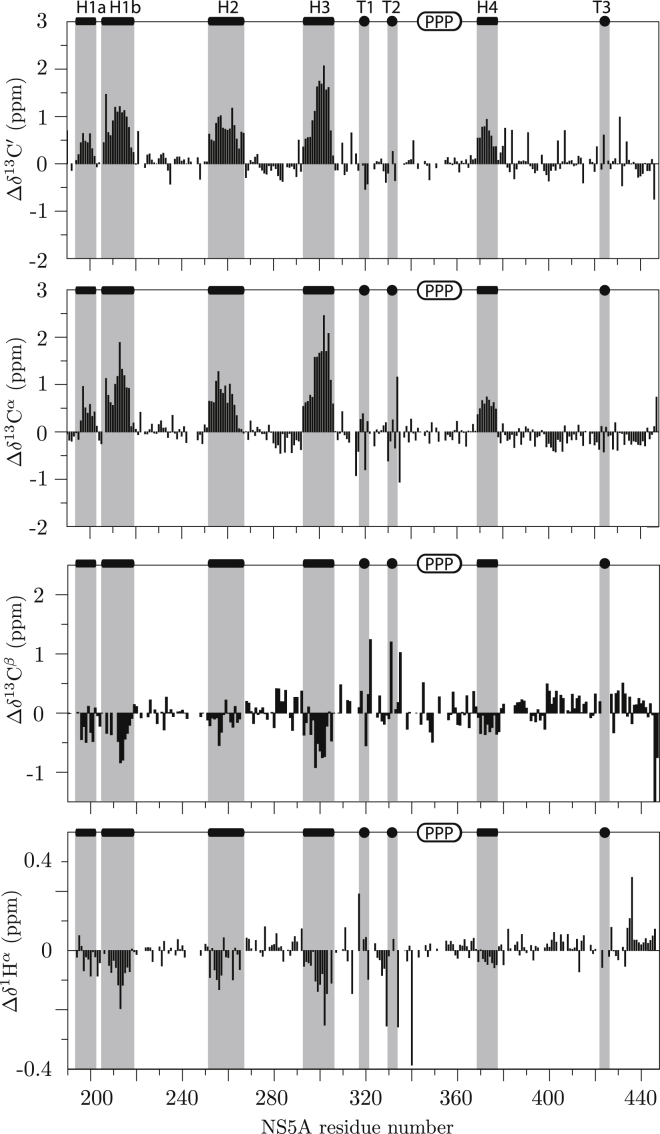
Secondary chemical shifts, obtained as the difference between the measured chemical shifts and computed residue-specific random coil (RC) values, present a first valuable tool to identify transient secondary structure along the peptide chain of intrinsically disordered proteins. Different RC shift predictors are available, and a comparison of 13Cα secondary chemical shifts calculated for NS5A-D2D3 using various predictors is shown in Fig. S3. The protocol of Kjaergaard et al. (52) and Kjaergaard and Poulsen (53) was chosen for further analysis, as these RC values resulted in the smallest deviation from zero for peptide regions without significant secondary structural propensity. The calculated secondary chemical shifts for 13C′, 13Cα, 13Cβ, and 1Hα of NS5A-D2D3 are plotted in Fig. 3. From these data, we can identify four peptide regions showing the characteristic signature for transiently structured α-helices: H1 (195–220), H2 (251–266), H3 (292–306), and H4 (370–378). In addition, H1 contains a proline residue that breaks the helix in two parts, H1a and H1b, as can be seen from the vanishing secondary chemical shift values close to this residue. As often observed for transiently formed α-helices in IDPs (54), helical regions H1b and H3 are N-capped by either an aspartate or a serine residue. In addition, the calculated Cα secondary chemical shift profile suggests that also a shorter H3 helix, capped by S297, is formed. Secondary structural propensity values, calculated using the NCSCP program (55), are shown in Fig. S4 a. The maximum helical propensities are 20% for H4, 35% for H2, 40% for H1, and 60% for H3. Helical wheel representations (Fig. S4 b) show that H1b, H2, and H3 are mainly made up of polar residues, while H1a and H4 are more hydrophobic. Another interesting observation is that H1b and H3 contain stretches of positively charged residues. The NS5A region comprising residues 315–334 also displays a particular secondary chemical shift pattern (Fig. 3) that is neither characteristic for α-helical nor for β-strand conformation, an observation that has previously been made by Rosnoblet et al. (29) for NS5A of HCV genotype 2a. Interestingly, this region is well conserved between different HCV genotypes (Fig. S7). For further analysis of the residual structure present in this functionally important region, we analyzed the measured chemical shifts with the MICS program (http://spin.niddk.nih.gov/bax/software/MICS/) (56) (Fig. S4 c). This algorithm predicts two type-I β-turns in this particular region, made up of residues 318–321 (T1) and 331–334 (T2). An additional type-II β-turn is located in D3, residues 422–425 (T3). The amino-acid composition of these peptide regions corresponds to what is expected for β-turns, with a proline in position 2 for type-I turns, and a glycine in position 3 for type-II turns.
Figure 3.
13C and 1Hα secondary chemical shifts measured for NS5A-D2D3. The random coil shifts were calculated according to the protocol of Kjaergaard et al. (52) and Kjaergaard and Poulsen (53). All transiently formed secondary structures, identified from these data, are indicated on top of the graph, and highlighted (gray bars).
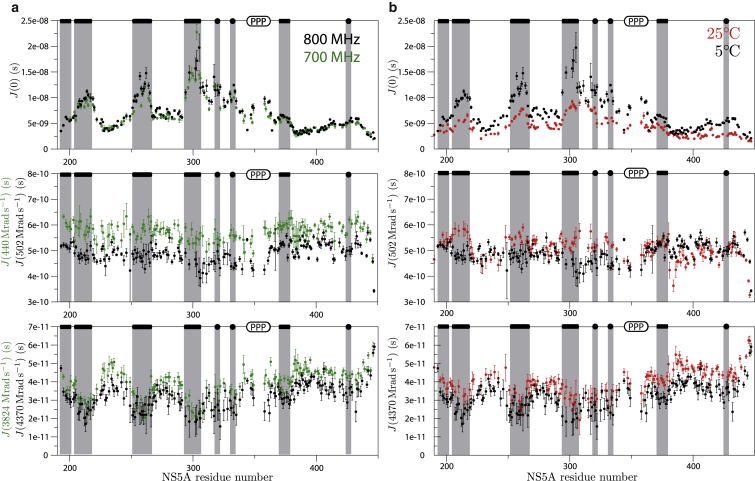
Additional site-resolved information on the conformational dynamics along the polypeptide chain has been obtained from 15N relaxation data. The measured 15N T1, T2, and the HetNOE data for each site (residue) can be converted into power spectral densities (46) at zero-frequency J(0), low frequency (J(ωN)), and high-frequency (J(0.87 ωH)) that are plotted in Fig. 4 as a function of the peptide sequence. In the following, we will focus on the J(0) values that are mainly affected by the slow overall and segmental tumbling motions present in the protein. As expected, higher J(0) values are observed for the transient secondary structural regions, providing an independent validation of our results derived from the analysis of secondary chemical shifts. Furthermore, J(0) values measured for regions with increased rigidity show a more pronounced change as a function of the sample temperature than residues in regions without residual structure. Because the 13C secondary chemical shifts indicate only a little change in helical propensity (37), this observation is most likely explained by a change in the solvent viscosity that slows down the segmental tumbling motions. Interestingly, such a strong J(0) temperature dependence is also observed for part of domain D3, indicating some increased rigidity in this highly flexible C-terminal protein region (residues 408–438). This observation may be explained by the presence of several hydrophobic residues (Y413, W433, and A436), which may form a transient hydrophobic cluster or be involved in additional intramolecular interactions. The measured J(0) values are independent of the magnetic field strength, indicating that conformational averaging occurs on a submicrosecond timescale, as slower timescale conformational exchange dynamics (μs-ms) are expected to give rise to magnetic-field-dependent line broadening, that are reflected in J(0) contributions.
Figure 4.
Power spectral densities extracted by reduced power spectral density mapping from 15N relaxation data (T1, T2, HetNOE) of NS5A-D2D3. (a) The J(0), J(ωN), and J(0.87ωH) values obtained at magnetic field strengths corresponding to 1H frequencies of 800 MHz (black) and 700 MHz (green). (b) Temperature dependence of power spectral densities measured at 25°C (red) and at 5°C (black) at 800 MHz. Transient secondary structures are indicated on top of the graph and highlighted (gray bars). To see this figure in color, go online.
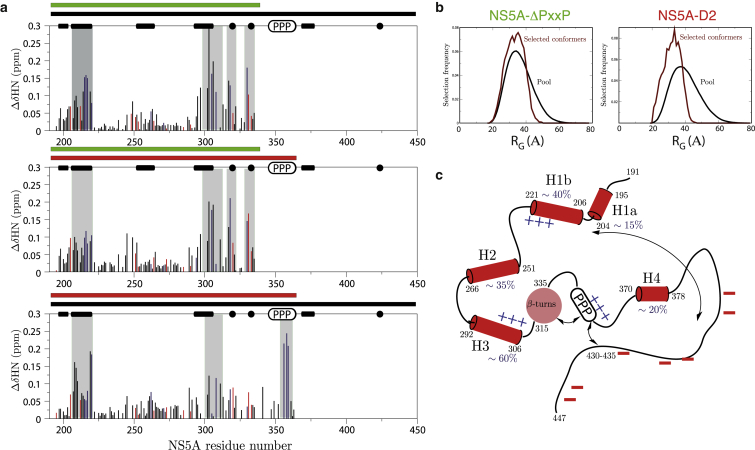
The method of choice to identify transient long-range interactions in IDPs by NMR is arguably the measurement of paramagnetic relaxation enhancement effects (PREs), a method that requires the introduction of a series of nitroxide spin labels at different natural or artificially created (by mutagenesis) cysteine positions in the protein. In 2012, using this approach, we had shown the presence of transient long-range contacts in the NS5A-D2 construct leading to a slight compaction of the protein’s structural ensemble. This result from PRE measurements was independently confirmed by SAXS data (37). A possible drawback of the PRE approach is that the mutations and/or the nitroxide spin labels may perturb the conformational dynamics of the wild-type protein. Here we opted for a different approach to obtain information on long-range interactions in NS5A-D2D3: the comparison of chemical shifts between different NS5A constructs under identical sample conditions. 1H, 15N chemical shift differences measured for pairs of NS5A constructs are plotted in Fig. 5 a. Chemical shift changes observed for residues that are not sequentially close to the truncation site are indicative of transient long-range interactions with the protein part that is absent in the shorter of the two constructs. Several such long-range interactions are clearly visible in the data of Fig. 5 a. For example, removing domain D3 has an impact on specific regions in domain D2, notably the helical regions H1b and H3, but also some residues in the polyproline-rich region. Interestingly, the peptide regions affected by the absence/presence of D3 all contain patches of positively charged residues. Because D3 is almost exclusively negatively charged, the observed chemical shift changes are most likely explained by a network of transient electrostatic interactions between the negatively charged D3 domain and positive charges in D2 and the polyproline region. Similar, transient contacts between the PxxP motif and the type-I β-turn region are observed. A comparison of 15N relaxation data (Fig. S5 a) and 13C secondary chemical shifts (Fig. S5 b) shows that the secondary structure, e.g., helical propensities, and local segmental conformational dynamics of NS5A-D2D3, are not significantly altered by these long-range interactions. Independent evidence of a slight compaction of the structural ensemble in the longer constructs is obtained from SAXS data (Fig. 5 b). EOM analysis (47) of the SAXS data obtained for two of the NS5A constructs, NS5A-D2 and NS5A-ΔPxxP, shows the radii of gyration distribution of the protein construct in comparison to that of a random coil chain of equal length. While the NS5A-ΔPxxP construct behaves in a manner similar to that of a random coil chain, NS5A-D2 clearly shows some tendency to favor more compact structures. Unfortunately, no exploitable SAXS data were obtained for NS5A-D2D3.
Figure 5.
Comparison of different NS5A constructs. (a) Chemical shift differences measured between pairs of NS5A constructs. The constructs used are indicated with bars at the top of each graph: NS5A-D2D3 (black bars), NS5A-D2 (red bars), and NS5A-ΔPxxP (green bars). Calculated chemical shift differences (vertical bars) plotted and color-coded according to the charge of the corresponding residues (blue, positively charged; red, negatively charged). Peptide regions that display significant chemical shift changes are highlighted (gray bars). (b) EOM (47) ensemble distributions calculated from SAXS data of the NS5A-D2 and NS5A-ΔPxxP constructs. The radius of gyration distribution of a pool of conformers (black), and of the EOM-selected conformers that best fit the experimental data (brown) is plotted. (c) Graphical sketch of the electrostatic interactions observed within the intrinsically disordered region of NS5A. To see this figure in color, go online.
The electrostatic nature of the long-range interactions has been confirmed by comparing NMR observables measured for NS5A-D2D3 under sample conditions differing in the ionic strength. 1H, 15N chemical shift changes and 15N T2 values, measured at two different salt concentrations, are shown in Fig. S5, c and d, respectively. These data show that the regions identified here to be involved in transient long-range interactions also show a more pronounced salt dependence, both in terms of structure (probed by chemical shift) and dynamics (probed by 15N relaxation). A graphical sketch highlighting all the local and long-range structural features of NS5A-D2D3 is shown in Fig. 5 c. In the following sections, we will address the question whether and, if yes, how these structural features are influenced by molecular interactions and phosphorylation.
NS5A-D2D3 structural changes induced by Bin1-SH3 binding
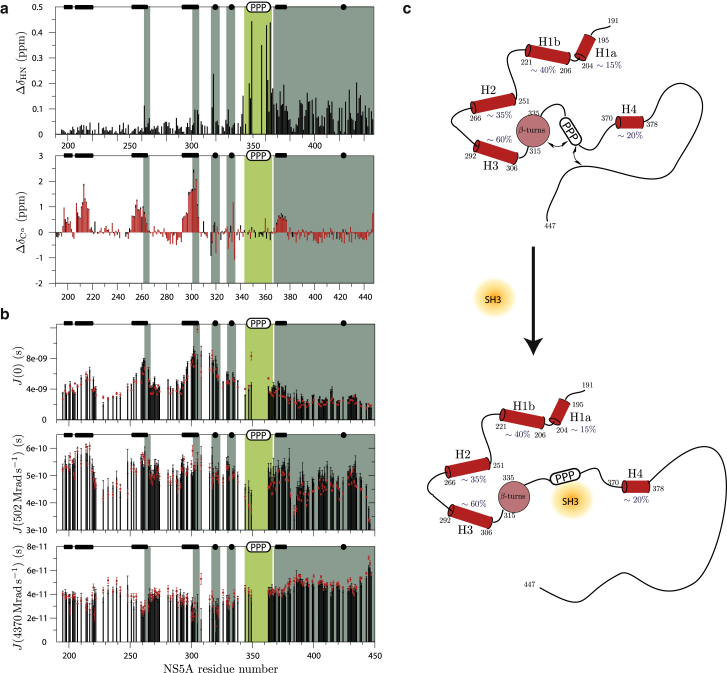
The tumor suppressor Bin1 interacts with NS5A via its SH3 domain. This interaction is suspected to play a role in the development of hepatocellular carcinoma in HCV-infected liver cells. In particular, it has been shown that NS5A is able to competitively displace the oncogenic Myc protein from its complex formed with Bin1-SH3 (57). Previously, we have found (37,38) that NS5A displays a high-affinity binding site (PxxP region), as well as two low-affinity binding sites (transient helices H1b and H3) for Bin1-SH3. The mode of interaction between Bin1-SH3 and NS5A-D2D3 was investigated by NMR spectroscopy. Chemical shift (and intensity) changes observed in the 1H-15N spectrum of NS5A-D2D3 (Fig. 6 a) after adding increasing amounts of unlabeled Bin1-SH3 confirms that the exchange between free and bound states is slow on the NMR timescale (high-affinity binding), and that the binding site is located in the polyproline-rich region of NS5A, as previously reported for the shorter NS5A-D2 construct (37). Surprisingly, the observed chemical shift changes are not limited to residues around the binding site (PxxP motif), but also observed for nearly the entire domain D3, as well as residues in H2, H3, and the type-I β-turn region. Additional NMR data recorded on a 1:1 NS5A-D2D3:Bin1-SH3 complex indicate that Bin1-SH3 binding does not alter the secondary structural propensities, as seen from 13C secondary chemical shifts (Fig. 6 a), but results in increased conformational flexibility, especially in domain D3, as seen from 15N relaxation data (Fig. 6 b). Size-exclusion chromatography data (Fig. S6), showing a larger hydrodynamic radius of the structural ensemble in the complex compared to the free NS5A-D2D3, are in agreement with these NMR-derived conclusions.
Figure 6.
Interaction of NS5A-D2D3 with Bin1-SH3. (a) 1H-15N, and 13Cα-chemical shift differences measured between NS5A-D2D3 in its free form and a 1:1 complex with Bin1-SH3. (b) Power spectral densities measured for free NS5A-D2D3 (black) and in complex with Bin1-SH3 (red). (c) Graphical sketch of the conformational change induced by SH3 binding. To see this figure in color, go online.
Taken together, our experimental data suggest the following model for the conformational changes induced by the interaction of the Bin1-SH3 domain with the intrinsically disordered part of NS5A (Fig. 6 c): the SH3 domain binds to the PxxP motif, thus shielding the positive charges of four consecutive positively charged residues (359–362) in the binding pocket; this changes the network of electrostatic long-range interaction in NS5A-D2D3, and results in higher flexibility, and better accessibility of D3.
CK2-phosphorylated NS5A-D2D3: phosphorylation sites, kinetics, and conformational changes
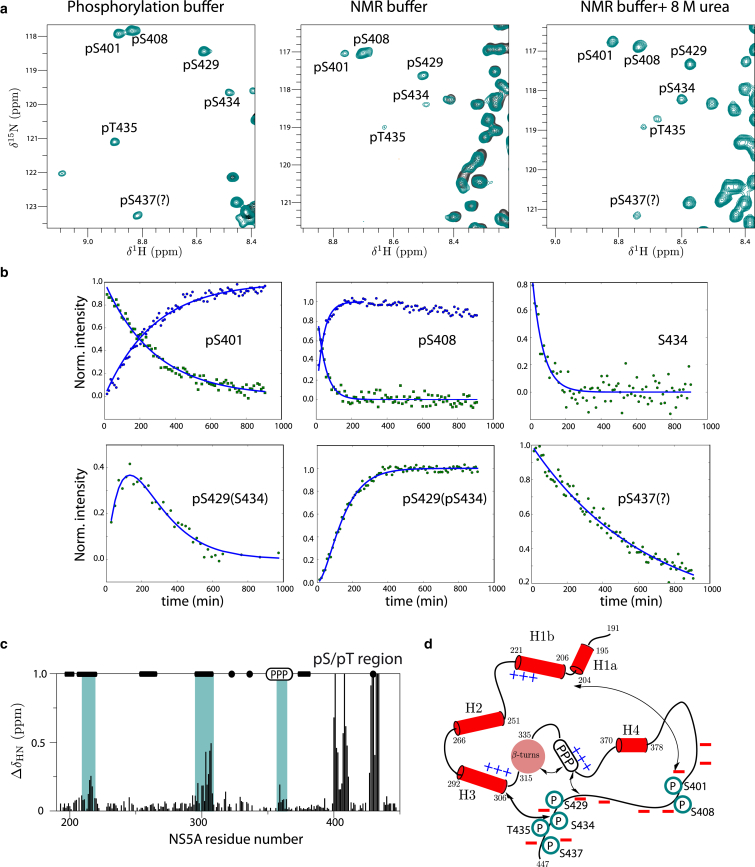
Heteronuclear NMR spectroscopy is a particularly powerful tool for obtaining site-resolved information on the sites, kinetics, and sequentiality of phosphorylation events in proteins, as well as for characterizing local changes in the conformational properties induced by the phosphorylation. We have thus followed in vitro phosphorylation of NS5A in real-time by recording a series of 1H-15N spectra during the phosphorylation reaction. This has allowed us to detect the presence of seven phosphorylated NS5A sites in D3, five of which (S401, S408, S429, S434, and T435) could be unambiguously assigned to specific NS5A residues, and an additional one was tentatively assigned to S437 (Fig. 7 a). More details about the NMR assignment of the phosphorylated residues, and the extraction of kinetic rate constants for the different phosphorylation events, will be given in the following sections.
Figure 7.
Phosphorylation study of NS5A-D2D3 by CK2. (a) 1H-15N correlation spectra of NS5A-D2D3 (green) measured 380 min after adding CK2 kinase. The spectral region, displaying cross peaks of phosphoserine and phosphothreonine residues (annotated in the spectra), is shown for the sample in the phosphorylation buffer, in the NMR buffer, and after adding 8 M urea. (Black) Reference spectrum before phosphorylation. (b) Kinetics traces (normalized peak intensity plotted as a function of reaction time) are shown for representative cross peaks of residues directly involved in phosphorylation, or close to a phosphorylation site. The kinetic data were fitted either to a model assuming a single phosphorylation event (and therefore single-exponential kinetics), or (in the case of intermediates, i.e., intensity buildup followed by decay) to a model of two consecutive phosphorylation events. (Lines) Results of the fitting. (c) 1H-15N chemical shift changes in NS5A-D2D3 upon phosphorylation by CK2 indicating structural changes in remote regions (indicated by green bars). (d) Model of the long-range electrostatic interactions with negative charges introduced by the phosphorylation of residues in D3. To see this figure in color, go online.
A first idea about the phosphorylated sites was obtained from spectral changes observed during the phosphorylation kinetics. However, because of the sequential proximity of several serine and threonine residues in D3, simple inspection of these data was not sufficient for unambiguous assignment. Therefore, the phosphorylation reaction was stopped after 380 min, and the sample was dialyzed back to the standard NMR buffer for which the NMR assignments are known. With the help of a few three-dimensional BT-HNC correlation experiments, this allowed unambiguous assignment of the four phosphoserines pS401, pS408, pS429, and pS434 (Fig. 7 a). The remaining correlation peaks in the spectral region typical for phosphoserines and phosphothreonines were very weak, making it difficult to detect correlations to neighboring residues. In order to remove hydrophobic interactions and residual structure that could explain the observed line broadening, we added 8 M urea to the sample. Under this denaturing condition, a better signal/noise in this spectral region was obtained (Fig. 7 a), and we could confirm our previous assignments and identify pT435 as an additional phosphorylated site. A last correlation peak was tentatively assigned to pS437, based on perturbations of neighboring residues. The corresponding site in NS5A of HCV genotype 2a was also shown to be phosphorylated by CK2, where it gives a phenotypic response to mutagenesis (27).
Kinetic traces (peak intensity as a function of time) can be extracted for all correlation peaks that are affected by the phosphorylation event(s). Selected examples of such kinetic curves are shown in Fig. 7 b. In the case of a single phosphorylation event, we expect these kinetic curves (intensity buildups or decays) to be monoexponential and characterized by a single time constant. In this context, we have multiple phosphorylation events of close-by NS5A D3 residues, representing competitive binding sites for the kinase. As a consequence, extracting kinetic rates is more complicated and requires certain assumptions. For pS401 and pS408, as well as the tentatively assigned pS437, we assume that the corresponding kinetic traces are only affected by a single phosphorylation process, and can thus be fitted by a monoexponential function. For pS429, pS434, and pT435, a first analysis indicates that the three sites are phosphorylated at similar rates. However, we could also detect a correlation peak that builds up first, and then decays again, characteristic of an intermediate species (Fig. 7 b). This peak could be assigned to a state where S429 is phosphorylated, but not S434 or T435. This intermediate peak kinetics fits well to a model of consecutive kinetics, where S429 is phosphorylated first, followed by phosphorylation of S434/T435 (considered a single event). Similar intermediate kinetics, detected for other correlation peaks (S432, W433, and D428) that are affected by the same events, could be fitted to the same kinetic model. This indicates that CK2 has a higher preference to phosphorylate S429 than S434 and T435. A summary of the kinetic rates and sequentiality of phosphorylation events is given in Table 1.
Table 1.
Kinetic rate constants
| Order of Phosphorylation | Kinetic Rate Constant (s−1) |
|---|---|
| pS408 | 1.59 ± 0.23 |
| pS429 | 0.76 ± 0.03 |
| pS434/pT435 | 0.68 ± 0.22 |
| pS401 | 0.22 ± 0.02 |
| pS437 | 0.09 ± 0.01 |
Kinetic rate constants were measured for the identified CK2 phosphorylation sites in NS5A-D2D3, assuming independent phosphorylation events for S401 and S408, and therefore fitting the decay as well as the buildup curves to monoexponential functions. In the case of residues S434, T435, and S437, the rates were extracted from fitting the cross-peak intensities of residues neighboring the phosphorylation sites. Errors were calculated from standard deviations of the fitted kinetic rate constants for resolved peaks affected by the same phosphorylation event (belonging to the same species).
To confirm our results also in the context of full-length NS5A, we performed additional in vitro CK2 phosphorylation NMR experiments on NS5A-FL. The sequentiality of the events and the phosphorylation sites were found to be the same as identified for NS5A-D2D3. The full-length protein, however, is even more prone to proteolytic degradation than the NS5A-D2D3 construct, which made a more detailed analysis impossible. Nevertheless, our NMR data shows that the phosphorylation of NS5A D3 residues by CK2 is independent of the presence of D1 and the N-terminal amphipathic helix.
Finally, we were interested in changes of the NS5A structural ensemble induced by the phosphorylation of several residues in D3. As usual, 1H and 15N chemical shift differences between phosphorylated and unphosphorylated NS5A-D2D3 were used to identify remote effects along the NS5A sequence (Fig. 7 c). Such remote chemical shift changes were observed for the transient helices H1b and H3, as well as the polyproline region, which all contain clusters of positively charged residues. Again, electrostatic interactions between remote peptide fragments of opposite net charge seem to be responsible for the observed effects. The addition of negative charges in D3 most likely enhances the population of more compact structures, or alternatively, changes the exchange kinetics. A change in the exchange kinetics may also explain the line broadening observed for pS434 and pT435, as well as some neighboring residues, upon CK2 phosphorylation.
Discussion
Intrinsic structural disorder is a common feature in many eukaryotic and viral proteins that are involved in regulatory and signaling processes, where transient binding to multiple partners (that are often regulated by posttranslational modifications, e.g., phosphorylation) plays an important role. Here we have studied the C-terminal, intrinsically disordered part, of NS5A from the hepatitis C virus, a multifunctional protein that is crucial for viral replication and particle assembly. Using NAPols as stabilizing agents, we were able to record, to our knowledge, the first NMR spectra of full-length NS5A that were of good quality. Comparison of the NMR spectra recorded on full-length NS5A, and NS5A-D2D3 (a construct that comprises the entire disordered domain), showed that the conformational properties of this C-terminal part in the full-length protein are similar to those in the isolated fragment. No significant interaction between the structured D1 domain with either D2 or D3 was observed, making the NS5A-D2D3 construct a valid model system to investigate transient structure in this NS5A region, as well as biologically relevant interactions with partners, and phosphorylation-induced changes in structure and dynamics.
Although the signal dispersion in the NMR spectra of NS5A-D2D3 is characteristic for an IDP, we have shown that there is a wealth of transient secondary and tertiary structure present in this highly flexible C-terminal part of NS5A. In particular, we have identified four peptide regions with significant helical propensities, three of them in D2 and one in D3. So far, no functional role has been ascribed to the peptide regions forming these transient helical structures, but often helical structures in IDPs are molecular recognition elements for the binding to partners (58). In addition to the four transient helical regions, we have also identified two type-I β-turns in a functionally important region (315–334) that is conserved between different genotypes (Fig. S7), and that interacts with the viral NS5B protein and CypA (29). Single-alanine mutations (P314A, P324A, S328A, and W329A) and a triple-alanine mutation (326–328) in this NS5A region have been shown to be lethal for the virus, resulting in replication-incapable mutants, while A317G and Y321A mutations result in severely reduced replication and a small-colony phenotype (27). It remains to be elucidated whether these particular β-turn structures are important for intermolecular interactions, and thus for viral replication and/or particle assembly. However, we may speculate that CypA has a role in stabilizing or destabilizing the β-turn structures by catalyzing the isomerization of the proline residue that has to be in trans-conformation for type I and II β-turn formation. We could further demonstrate some local rigidity in the otherwise highly dynamic domain D3. We may speculate that the observed increased rigidity is due to the presence of a type-II β-turn favoring more compact structures in the conformational ensemble, and/or hydrophobic clustering of aromatic side chains present in this region of NS5A. Again, it remains to be investigated whether these structural features play a role in the interaction of D3 with the HCV core protein, or with host factors such as lysine methyltransferase SET- and MYND-domain-containing protein 3 (SMYD-3), identified in 2014 (59).
A major focus of our NMR study was the investigation of the presence and nature of transient long-range interactions in NS5A-D2D3, and their modulation by binding to other proteins or by phosphorylation. We could experimentally demonstrate the presence of a network of electrostatic interactions between peptide segments that are characterized by a clustering of either positive (in D2 and the polyproline-rich region) or negative (in D3) charges. Such interactions have been shown theoretically and experimentally to dominate the dimensions of the conformational ensemble of IDPs (60,61). In particular, molecular dynamics simulations have shown that the clustering of charged residues has a strong impact on the hydrodynamic radius of the IDP. The long-range interactions observed in the intrinsically disordered region of NS5A are an example for such electrostatically driven interdomain-interactions, resulting in a compaction of the structural ensemble. The patches of positively and negatively charged residues in NS5A-D2D3 are well conserved among the HCV genotypes (see Fig. S7), suggesting that they may be functionally important. Mutational studies are required to further elucidate the role of these electrostatic long-range interactions during different stages in the viral life cycle. We could also show that these long-range interactions, and thus the overall compactness of the NS5A protein, can be modulated by binding of a SH3 domain to the PxxP motif, masking a patch of positive charges, or by introducing additional negative charges via phosphorylation of serine and threonine side chains. The modulation of long-range interactions may be involved in the regulatory mechanisms that switch NS5A between its different functions in HCV-infected cells.
Finally, our study allowed the identification of several, so-far-unreported, CK2 phosphorylation sites in domain D3 of NS5A from HCV genotype 1b, and provided some information about the preference of the kinase for the different phosphorylation sites in D3. Two of the identified phosphorylation sites (S408 and T435) are located in peptide regions with sequences that fit the consensus recognition motif T/SXXD/E, while others (S401, S429, and S434) do not fit this simple rule. However, if there is no negatively charged residue at position i+3, a negatively charged residue at position i+1 also becomes a CK2 site. Consequently, S408 has been found in our study to be phosphorylated at the highest rate, as it is part of a canonical CK2 recognition site that is freely accessible to the kinase. Residues S429 and S434 and S435 are phosphorylated at similar rates. Most likely, phosphorylation of S429 is required to increase the kinase affinity for the remaining sites (S434/S435), either by the additional negative charge or by a change in local structure. Similarly, the negative charges of pS434 and pS435 will favor phosphorylation of S437. Interestingly, the major CK2 phosphorylation site S437, with a functional role in particle assembly in genotype 2a (27), was found in our study to be only a minor site that becomes phosphorylated at a much slower rate. This finding can be explained by the fact that the T/SXXD/E motif, forming an ideal substrate for CK2, is not conserved in NS5A of genotype 1b (SEEA). Our results are, however, in agreement with the observations of Masaki et al. (35) in 2014, who found that among the three serines in the cluster S432-S434-S437, phosphomimetic mutations of at least two of them are required in order to restore viral particle production, and interaction of NS5A with the viral core protein. These observations point toward the conclusion that the overall distribution of negative charges in this particular D3 region is more important for the function of the protein than the exact position of the phosphorylated residues that varies among different genotypes. This finding is also in agreement with our hypothesis that an important role of CK2 phosphorylation is the modulation of long-range electrostatic interactions in NS5A.
Author Contributions
D.W. and B.B. designed the research; Z.S., P.M., and M.S. performed research and data analysis; M.B., A.P., and G.D. synthesized NAPol; and Z.S. and B.B. wrote the article with the help of all co-authors.
Acknowledgments
The authors thank Silke Hoffmann (Forschungszentrum Jülich), Phil Selenko, and François Theillet (Forschungsinstitut für Molekulare Pharmakologie Berlin) for stimulating discussion on NS5A and protein phosphorylation, and Adrien Favier and Isabel Ayala (Institut de Biologie Structurale Grenoble) for technical support and help with protein expression.
We acknowledge financial support by the Commissariat à l’Énergie Atomique et aux Énergies Alternatives (CEA), the Centre National de la Recherche Scientifique (CNRS), the University Grenoble Alpes, the Deutsche Forschungsgemeinschaft (DFG grant No. SFB974), the Agence Nationale de la Recherche (ANR grant No. X-Or-2010-BLAN-1535), and the European Commission (grant No. FP7-MC-ITN-IDP, NMR contract No. 264257). This work used the platforms of the Grenoble Instruct Center (ISBG grant No. UMS 3518 CNRS-CEA-UJF-EMBL) with support from the French Infrastructure for Integrated Structural Biology (grant No. ANR-10-INSB-05-02) and New Generation of Drugs for Alzheimer’s Disease (GRAL grant No. ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. Financial support from the Très Grande Infrastructure de Recherche: Résonance Magnétique Nucléaire à Très Hauts Champs (TGIR-RMN-THC grant No. FR3050, CNRS for NMR Measurements on NS5A-FL) is also gratefully acknowledged.
Editor: H. Jane Dyson.
Footnotes
Seven figures and a sequence alignment of NS5A from different HCV genotypes are available at http://www.biophysj.org/biophysj/S0006-3495(15)00623-2.
Supporting Material
References
- 1.Communie G., Ruigrok R.W., Blackledge M. Intrinsically disordered proteins implicated in paramyxoviral replication machinery. Curr. Opin. Virol. 2014;5:72–81. doi: 10.1016/j.coviro.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Davey N.E., Travé G., Gibson T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue B., Blocquel D., Longhi S. Structural disorder in viral proteins. Chem. Rev. 2014;114:6880–6911. doi: 10.1021/cr4005692. [DOI] [PubMed] [Google Scholar]
- 5.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Dunker A.K., Oldfield C.J., Uversky V.N. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J., Thelen J.J., Xu D. MUSITE, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol. Cell. Proteomics. 2010;9:2586–2600. doi: 10.1074/mcp.M110.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 9.Theillet F.X., Smet-Nocca C., Selenko P. Cell signaling, post-translational protein modifications and NMR spectroscopy. J. Biomol. NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoemaker B.A., Portman J.J., Wolynes P.G. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X., Xue B., Uversky V.N. The intrinsic disorder status of the human hepatitis C virus proteome. Mol. Biosyst. 2014;10:1345–1363. doi: 10.1039/c4mb00027g. [DOI] [PubMed] [Google Scholar]
- 12.Nanda S.K., Herion D., Liang T.J. The SH3 binding motif of HCV [corrected] NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology. 2006;130:794–809. doi: 10.1053/j.gastro.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Tan S.L., Nakao H., Katze M.G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosztányi Z., Csizmok V., Simon I. IUPRED: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 15.Penin F., Brass V., Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 16.Tellinghuisen T.L., Marcotrigiano J., Rice C.M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brass V., Bieck E., Moradpour D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 18.Tellinghuisen T.L., Marcotrigiano J., Rice C.M. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 2004;279:48576–48587. doi: 10.1074/jbc.M407787200. [DOI] [PubMed] [Google Scholar]
- 19.Love R.A., Brodsky O., Cronin C.N. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert S.M., Langley D.R., Matthews S.J. The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors. Protein Sci. 2014;23:723–734. doi: 10.1002/pro.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordek D.G., Croom-Perez T.J., Cameron C.E. Expanding the proteome of an RNA virus by phosphorylation of an intrinsically disordered viral protein. J. Biol. Chem. 2014;289:24397–24416. doi: 10.1074/jbc.M114.589911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang L., Hwang J., Cameron C.E. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 2005;280:36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 23.Gentzsch J., Brohm C., Pietschmann T. Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathog. 2013;9:e1003355. doi: 10.1371/journal.ppat.1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanoulle X., Badillo A., Lippens G. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept. Lett. 2010;17:1012–1018. doi: 10.2174/092986610791498920. [DOI] [PubMed] [Google Scholar]
- 25.Hanoulle X., Verdegem D., Lippens G. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 2009;381:634–638. doi: 10.1016/j.bbrc.2009.02.108. [DOI] [PubMed] [Google Scholar]
- 26.Tellinghuisen T.L., Foss K.L., Rice C.M. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 2008;82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tellinghuisen T.L., Foss K.L., Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rai D., Wang L., Liu X. The changing face of hepatitis C: recent advances on HCV inhibitors targeting NS5A. Curr. Med. Chem. 2015 doi: 10.2174/0929867322666150209150920. Published online February 9, 2015. [DOI] [PubMed] [Google Scholar]
- 29.Rosnoblet C., Fritzinger B., Hanoulle X. Hepatitis C virus NS5B and host cyclophilin A share a common binding site on NS5A. J. Biol. Chem. 2012;287:44249–44260. doi: 10.1074/jbc.M112.392209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross-Thriepland D., Harris M. Insights into the complexity and functionality of hepatitis C virus NS5A phosphorylation. J. Virol. 2014;88:1421–1432. doi: 10.1128/JVI.03017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintavalle M., Sambucini S., Neddermann P. Hepatitis C virus NS5A is a direct substrate of casein kinase I-α, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J. Biol. Chem. 2007;282:5536–5544. doi: 10.1074/jbc.M610486200. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y.C., Su W.C., Lai M.M.C. Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J. Virol. 2010;84:7983–7993. doi: 10.1128/JVI.00068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neddermann P., Quintavalle M., De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 2004;78:13306–13314. doi: 10.1128/JVI.78.23.13306-13314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu D., Lemm J.A., Fridell R.A. The effects of NS5A inhibitors on NS5A phosphorylation, polyprotein processing and localization. J. Gen. Virol. 2011;92:2502–2511. doi: 10.1099/vir.0.034801-0. [DOI] [PubMed] [Google Scholar]
- 35.Masaki T., Matsunaga S., Suzuki T. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production. J. Virol. 2014;88:7541–7555. doi: 10.1128/JVI.03170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Appel N., Zayas M., Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feuerstein S., Sólyom Z., Brutscher B. Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J. Mol. Biol. 2012;420:310–323. doi: 10.1016/j.jmb.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Schwarten M., Sólyom Z., Brutscher B. Interaction of nonstructural protein 5A of the hepatitis C virus with Src homology 3 domains using noncanonical binding sites. Biochemistry. 2013;52:6160–6168. doi: 10.1021/bi400363v. [DOI] [PubMed] [Google Scholar]
- 39.Bazzacco P., Billon-Denis E., Popot J.L. Nonionic homopolymeric amphipols: application to membrane protein folding, cell-free synthesis, and solution nuclear magnetic resonance. Biochemistry. 2012;51:1416–1430. doi: 10.1021/bi201862v. [DOI] [PubMed] [Google Scholar]
- 40.Sharma K.S., Durand G., Pucci B. Glucose-based amphiphilic telomers designed to keep membrane proteins soluble in aqueous solutions: synthesis and physicochemical characterization. Langmuir. 2008;24:13581–13590. doi: 10.1021/la8023056. [DOI] [PubMed] [Google Scholar]
- 41.Sharma K.S., Durand G., Pucci B. Non-ionic amphiphilic homopolymers: synthesis, solution properties, and biochemical validation. Langmuir. 2012;28:4625–4639. doi: 10.1021/la205026r. [DOI] [PubMed] [Google Scholar]
- 42.Favier A., Brutscher B. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR. 2011;49:9–15. doi: 10.1007/s10858-010-9461-5. [DOI] [PubMed] [Google Scholar]
- 43.Sólyom Z., Schwarten M., Brutscher B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F., Grzesiek S., Bax A. NMRPIPE: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 46.Farrow N.A., Zhang O., Kay L.E. Spectral density function mapping using 15N relaxation data exclusively. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 47.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 48.Glück J.M., Wittlich M., Koenig B.W. Integral membrane proteins in nanodiscs can be studied by solution NMR spectroscopy. J. Am. Chem. Soc. 2009;131:12060–12061. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 49.Feuerstein S., Plevin M.J., Brutscher B. iHADAMAC: a complementary tool for sequential resonance assignment of globular and highly disordered proteins. J. Magn. Reson. 2012;214:329–334. doi: 10.1016/j.jmr.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 50.Lescop E., Rasia R., Brutscher B. Hadamard amino-acid-type edited NMR experiment for fast protein resonance assignment. J. Am. Chem. Soc. 2008;130:5014–5015. doi: 10.1021/ja800914h. [DOI] [PubMed] [Google Scholar]
- 51.Feuerstein S., Solyom Z., Brutscher B. 1H, 13C, and 15N resonance assignment of a 179 residue fragment of hepatitis C virus non-structural protein 5A. Biomol. NMR Assign. 2011;5:241–243. doi: 10.1007/s12104-011-9309-2. [DOI] [PubMed] [Google Scholar]
- 52.Kjaergaard M., Brander S., Poulsen F.M. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 53.Kjaergaard M., Poulsen F.M. Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR. 2011;50:157–165. doi: 10.1007/s10858-011-9508-2. [DOI] [PubMed] [Google Scholar]
- 54.Jensen M.R., Zweckstetter M., Blackledge M. Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem. Rev. 2014;114:6632–6660. doi: 10.1021/cr400688u. [DOI] [PubMed] [Google Scholar]
- 55.Tamiola K., Acar B., Mulder F.A. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 56.Shen Y., Bax A. Identification of helix capping and b-turn motifs from NMR chemical shifts. J. Biomol. NMR. 2012;52:211–232. doi: 10.1007/s10858-012-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aladag A., Hoffmann S., Schwarten M. Hepatitis C virus NS5A is able to competitively displace c-Myc from the Bin1 SH3 domain in vitro. J. Pept. Sci. 2014;20:334–340. doi: 10.1002/psc.2618. [DOI] [PubMed] [Google Scholar]
- 58.Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberle C.A., Zayas M., Superti-Furga G. The lysine methyltransferase SMYD3 interacts with hepatitis C virus NS5A and is a negative regulator of viral particle production. Virology. 2014;462-463:34–41. doi: 10.1016/j.virol.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Müller-Späth S., Soranno A., Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.