Table 6. Core Modifications.
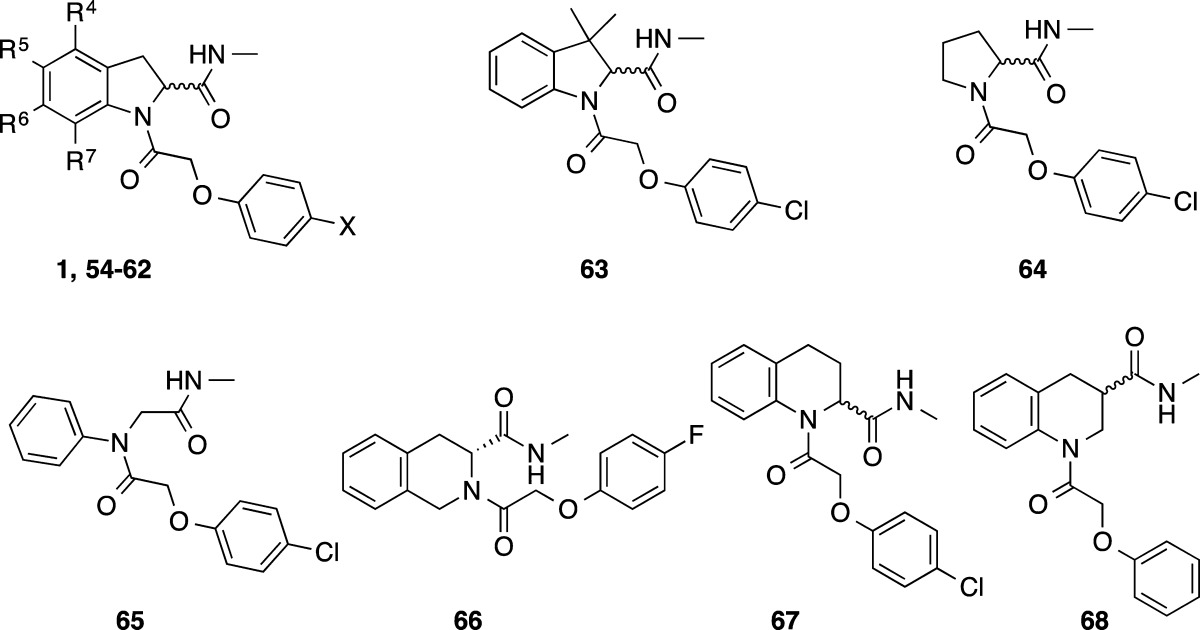
| microsomal
intrinsic clearance (mL min–1 g–1)b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| compd no. | stereochem | X | R4 | R5 | R6 | R7 | T. b. brucei EC50 (μM)a | MRC-5 EC50 (μM)a | m | r | h |
| 1 | R/S | Cl | H | H | H | H | 0.03 | >50 | 4.4 | 4.4 | 2.6 |
| 54 | R/S | Cl | H | F | H | H | 0.06 | >50 | 2.8 | 5.3 | 1.6 |
| 55 | R/S | F | H | F | H | H | 0.08 | >50 | 1.3 | 2.1 | 0.5 |
| 56 | R | F | H | F | H | H | 0.04 | >50 | ND | ND | ND |
| 57 | R/S | F | H | F | F | H | 0.09 | >50 | 2.0 | 2.8 | 1.9 |
| 58 | R/S | F | H | F | H | F | 0.08 | >50 | 2.9 | ND | ND |
| 59 | R/S | Cl | F | F | F | H | 0.07 | >50 | 3.7 | 6.7 | 4.3 |
| 60 | R/S | F | H | Br | H | H | 0.08 | >50 | 4.6 | ND | ND |
| 61 | R/S | F | H | Ph | H | H | 0.24 | >50 | ND | 25 | ND |
| 62 | R/S | F | H | 4-pyridyl | H | H | 0.44 | >50 | ND | ND | ND |
| 63 | R/S | Cl | NAc | NA | NA | NA | 0.92 | >50 | 14 | ND | ND |
| 64 | Cl | NA | NA | NA | NA | 36 | >50 | ND | ND | ND | |
| 65 | Cl | NA | NA | NA | NA | 19 | >50 | 4.7 | ND | ND | |
| 66 | R | F | NA | NA | NA | NA | 37 | >50 | ND | ND | ND |
| 67 | R/S | Cl | NA | NA | NA | NA | 0.46 | >50 | ND | ND | ND |
| 68 | R/S | NA | NA | NA | NA | >50 | >50 | ND | ND | ND | |
EC50 values are shown as mean values of two or more determinations. Standard deviation is typically within 2–3 fold from the EC50.
m/r/h = mouse/rat/human. Synthetic scheme outlined in Supporting Information,30. ND = not determined.
NA = not applicable.
