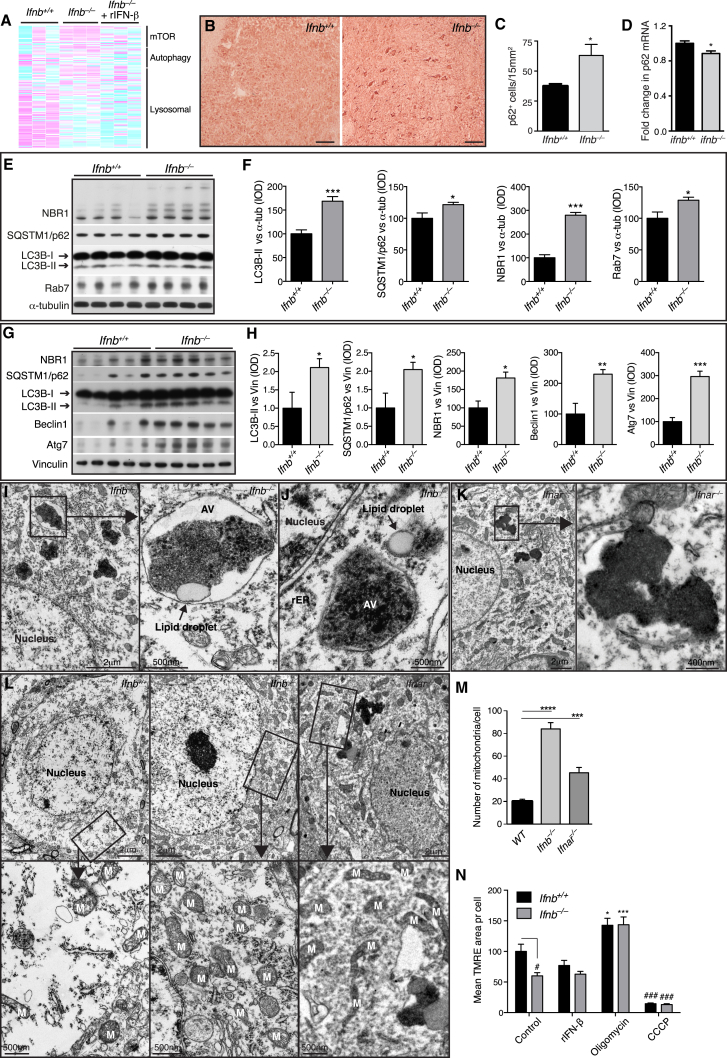
Figure 4.
Late-Stage Degradative Autophagosomes Accumulate in Ifnb–/– Neurons
(A) Heatmaps of GSEA core-enriched genes in autophagy-related pathways comparing Ifnb+/+ and Ifnb–/– mouse CGNs, Ifnb–/– with or without 24 hr rIFN-β (100 U/ml), n = 3.
(B) IHC from pons area using p62 antibodies. Scale bar, 20 μm.
(C) Quantification of p62+ cells/15 mm2. Data are mean ± SEM, n = 5.
(D) Fold change in p62 mRNA in brain extracts of 3-month-old mice. Data are mean ± SEM, n = 3.
(E–H) WB of BG of (E) 1.5-month-old and (G) 6- to 8-month-old mice and (F) and (H) quantified IOD of bands. Data are mean ± SEM of four or five brains.
(I–L) TEM of 9-month-old mice thalami. (I–J) Ifnb–/– mice showing perinuclear electron-dense late-stage autophagic vacuoles (AV), most surrounded by single lipid membrane and associated with lipid droplets. rER, rough endoplasmic reticulum. (K) Ifnar–/– mouse with similar electron-dense aggregates. (L) TEM of mitochondria in thalamic neuron cell body. M, mitochondria.
(M) Graph, mean ± SEM mitochondria per cell body of 12–16 thalamic neurons; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by one-way ANOVA with post hoc Dunnett’s multiple comparisons test.
(N) TMRE analysis of 21-day-old CNs with mean TMRE area per cell, n = 3. ∗Treatment effect, and #genotype effect.
For (B), (C), (F), (H), and (N), ∗/#p < 0.05, ∗∗p < 0.01, ∗∗∗/###p < 0.001 by unpaired Student’s t test. See also Figure S4 and Table S2.