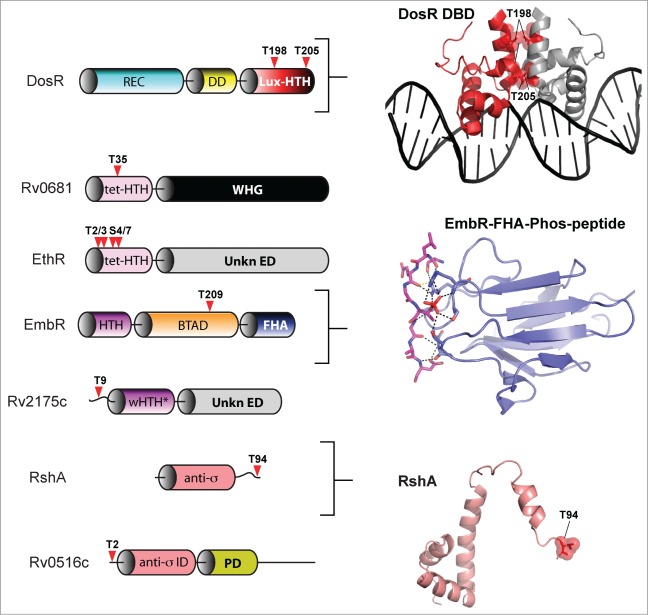
Figure 7.
eSTK phosphorylation sites in M. tuberculosis transcription factors. Annotation is as per Figure 4. Domain architectures are shown on the left, and 3-dimensional atomic structures and models on the right. The PDB codes for the DosR DNA-binding domain (DBD) and the EmbR-forkhead-associated domain (FHA; shown in blue) in complex with an interacting peptide (in magenta) and phosphate (shown in red) are 3C3W (ref. 163) and 2FF4 (ref. 114), respectively. The RshA anti σ-factor structure was generated with Swiss-Model using an available crystal structure from the anti-sigma factor ChrR (PDB code 2Q1Z; ref. 164) as a molecular template. eSTK phosphorylated residues are shown in red. Abbreviations not previous described are tet-HTH, TetR family helix-turn-helix domain; WHG, tryptophan-histidine-glycine domain presumed to bind a ligand or unknown effector molecule; UnKn ED, unknown effector domain; BTAD, bacterial transcriptional activator domain; wHTH*, wHTH domain missing helix 3; anti-σ: anti-sigma factor; anti-σ ID, anti-sigma factor interacting domain; PD, phosphorylation domain.