Figure 2.
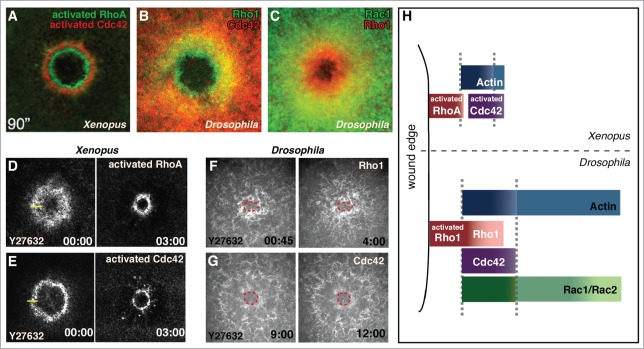
GTPase arrays form in response to cell wound repair the Xenopus oocyte and Drosophila syncytial embryo cell repair models. (A, D, and E) Fluorescent reporters for Xenopus studies use biosensor reporters depicting activated Rho family GTPases. (B, C, F, and G) Drosophila Rho GTPase fluorescent probes report all protein (not just the activated forms). (A) Activated RhoA and Cdc42 form non-overlapping concentric rings in the Xenopus oocyte (© 2005 Rockefeller University Press. Originally published in Journal of Cell Biology 168: 429–439). (B) Rho1 and Cdc42 form overlapping arrays in the Drosophila syncytial embryo with the Rho1 array interior to Cdc42. (C) Rac1 and Rho1 form overlapping arrays in the Drosophila syncytial embryo with the Rho1 array interior to Rac1. (D and E) RhoA and Cdc42 arrays in the Xenopus model translocate upon inhibition of contraction with the Rok inhibitor Y27632 (adapted from21 and reproduced with permission from Elsevier). (F and G) Rho1 and Cdc42 arrays in the Drosophila model are unable to translocate when contraction is inhibited with Y27632 (wound edge outlined in red). (H) Schematic depicting the relative localization of Rho family GTPases in the Xenopus and Drosophila single cell wound repair models (adapted from17 and reproduced with permission from Elsevier).
