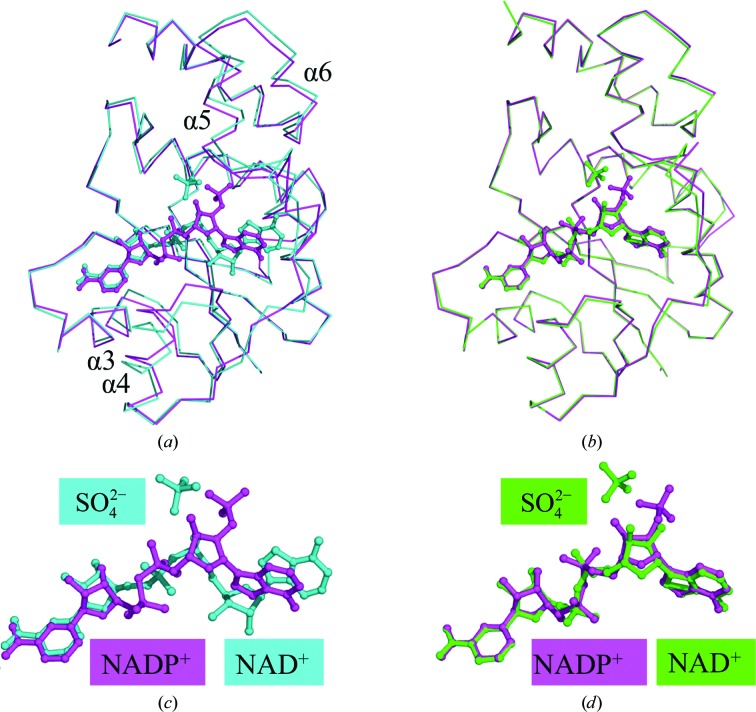
Figure 1.
(a) Superposition of ribbon diagrams of WT NMNAT complexed with NAD+ (cyan) compared with NADP+ (magenta). The largest deviation in WT NMNAT to accommodate NADP+ is seen in helices 4, 5 and 6. There is an overall opening of the active site to fit the larger NADP+ molecule. (b) Superposition of ribbon diagrams of His19Ala NMNAT complexed with NAD+ (green) compared with WT NMNAT with NADP+ (magenta). The NAD+ and NADP+ molecules are completely superposed in this alternative mode of dinucleotide binding to NMNAT. (c) Close-up of the superposition of NAD+ (cyan) and NADP+ (magenta) in WT NMNAT. The nicotinyl moiety is very closely aligned between NAD+ and NADP+, whereas the adenylyl moiety is completely misaligned. The adenylyl moiety has undergone a rotation to allow its adenine ring to interact with the side chain of Tyr126 of NMNAT. The ribose of the adenylyl moiety has also undergone a rotation to allow the 2′-phosphate to interact with His16 and His19 of NMNAT. (d) Close-up of the superposition of NAD+ in His19Ala NMNAT (green) and NADP+ in WT NMNAT (magenta). An identical rotation around the adenylyl moiety is observed between NAD+ and NADP+. A sulfate molecule mimicking the γ-phosphate of ATP was identified in the active sites of the WT and H19A NMNAT–NAD+ complexes and is shown for clarity.