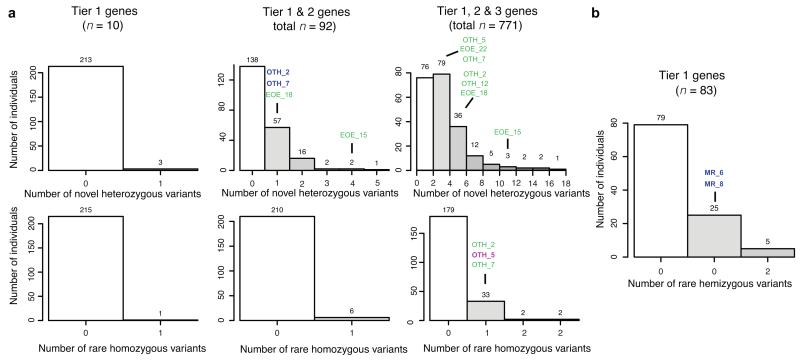
Figure 2. The burden of variants of unknown significance.
(a) Histograms of the number of previously unreported coding variants at conserved positions in different sets of candidate gene (Tiers 1, 1+2 and 1+2+3 for columns left to right) for early-onset epilepsy, under different inheritance models, across 216 WGS500 samples. (b) Histogram of the number of previously unreported coding variants at conserved positions in known X-linked mental retardation genes (XLMR), for the 99 male WGS500 samples. The candidate genes were chosen by high-throughput searches (Online Methods). Sample identifiers indicate individuals with the disorder in question. Sample names in green text indicate that the variant is not likely to be pathogenic (since it does not fit a plausible inheritance model or is less functionally compelling than another candidate); blue text indicates that the variant is thought to be causal (see Supplementary Table 6). OTH: Ohtahara syndrome; EOE: nonsyndromic early onset epilepsy; MR: mental retardation. See Supplementary Fig. 4 for the analysis of craniosynostosis.