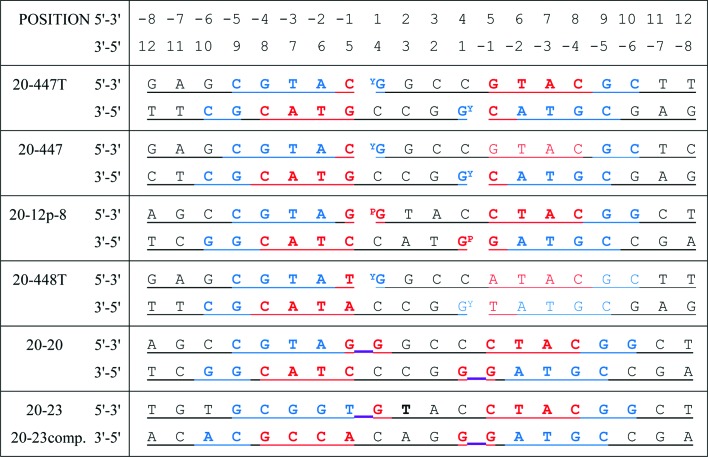
Figure 1.
DNA sequences used in successful crystallization experiments. DNAs are self-complementary and form 20-base-pair homoduplexes, except for 20-23, which forms a heteroduplex with 20-23comp (Miles et al., 2013 ▸). DNAs which are cleaved by the enzyme form a covalent bond with Tyr123 from GyrA (indicated by a superscript Y). The 20-12p-8 DNA duplex has an artificial nick in the DNA at each cleavage site, and the 5′ nucleotide of the 12-mer includes a 5′ phosphate (indicated by a superscript P). The 20-20 and 20-23 DNAs were not used in the crystals described in this paper, but as uncleaved DNA (the uncleaved link between the −1 and +1 nucleotides is underlined in purple) in crystal structures with the NBTIs GSK299423, GSK966587 and the Y123F mutant (PDB entries 2xcs, 2xcr and 4bul; Bax et al., 2010 ▸; Miles et al., 2013 ▸). Nucleotides coloured in red have their phosphates bound by GyrB and those in blue by GyrA. Note that the terminal bases are not always complementary. By convention, Topo2A DNA sequences are numbered relative to the cleavage sites (between −1 and 1).