Table 1. X-ray crystallographic data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| CA IX mimicsucrose | CA IX mimic | |
|---|---|---|
| PDB code | 4ywp | 4zao |
| Data-collection statistics | ||
| Temperature (K) | 100 | 100 |
| Wavelength () | 0.918 | 1.54 |
| Space group | P21 | P21 |
| Unit-cell parameters | ||
| a () | 41.8 | 41.8 |
| b () | 41.2 | 41.2 |
| c () | 72.3 | 72.0 |
| () | 103.8 | 103.8 |
| Reflections | ||
| Theoretical | 42836 | 22290 |
| Unique | 42193 | 21707 |
| Resolution () | 20.01.50 (1.501.45) | 20.01.80 (1.861.80) |
| R merge † (%) | 5.1 (10.3) | 4.5 (6.3) |
| I/(I) | 16.3 (11.7) | 17.3 (12.8) |
| Completeness (%) | 98.5 (99.7) | 97.5 (97.1) |
| Multiplicity | 3.7 (3.5) | 2.7 (2.7) |
| Model statistics | ||
| R cryst ‡ (%) | 18.0 | 17.5 |
| R free § (%) | 21.2 | 20.5 |
| Residue Nos. | 4261 | 4261 |
| No. of atoms¶ | ||
| Protein | 2132 | 2132 |
| Ligand | 27 | 0 |
| Water | 290 | 169 |
| R.m.s.d. | ||
| Bond lengths () | 0.009 | 0.006 |
| Bond angles () | 1.32 | 1.044 |
| Ramachandran statistics (%) | ||
| Favored | 89.0 | 94.8 |
| Allowed | 11.0 | 5.2 |
| Average B factors (2) | ||
| Main chain | 9.5 | 13.9 |
| Side chain | 13.4 | 18.6 |
| Ligand | 7.8 | |
| Solvent | 19.9 | 23.8 |
R
merge = 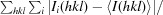
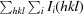 100.
100.
R
cryst = 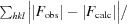
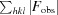 100.
100.
R free is calculated in the same manner as R cryst except that it uses 5% of the reflection data omitted from refinement.
Includes alternate conformations.
