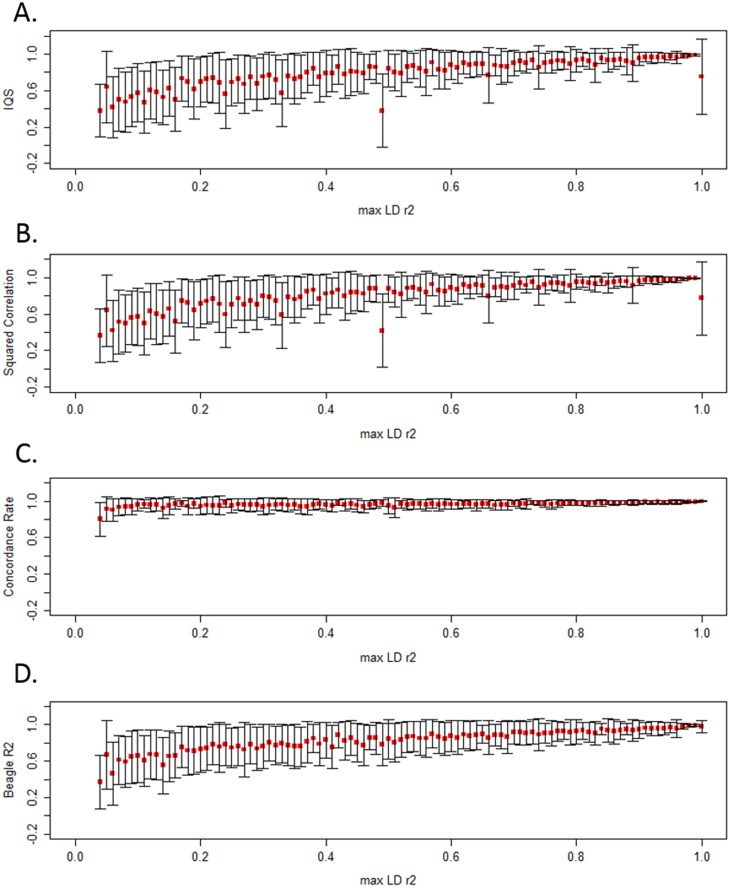
Fig 3. IQS, squared correlation, concordance rate, and BEAGLE R2 are shown in max r2 LD bins.
Mean accuracy of SNPs in each MAF bin (defined by 0.01 increments with N = 13,442 variants total) is denoted by the red dots and the bars indicate one standard deviation (above and below the mean). These results were produced by using the 1000 Genomes AFR reference population as the study sample with Omni 2.5M typed coverage on chromosome 15.