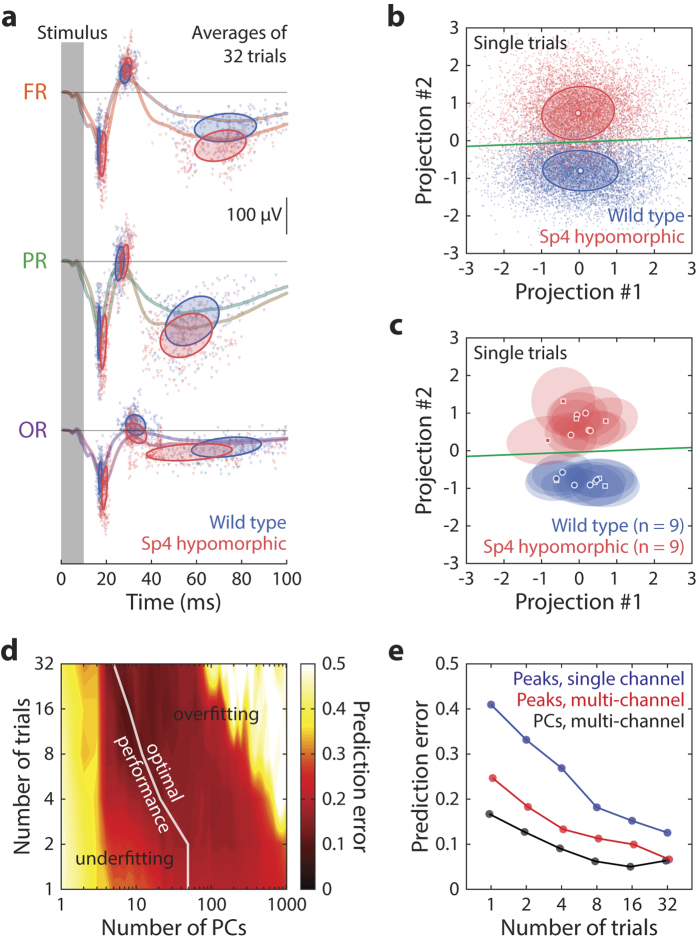
Figure 3. Characterizing time-domain ERP signatures unique to the mouse model under test.
(a) Illustration of ERP peak feature extraction used in traditional methods. Grand average ERPs from channels FR, PR and OR are plotted as colored curves, the grand average of wild type animals in blue and that of Sp4 hypomorphic mice in red. ERP peaks N1, P1, N2 detected from averages of 32-trial blocks are shown by triangular symbols, with upward-pointing triangles representing positive peaks and downward-pointing ones negative peaks. Ellipses mark the 1 σ boundaries of the covariances of the latency and magnitude of the detected peaks. (b) Linear projections of single-trial, full spatio-temporal ERPs (small, transparent dots) on to a plane, spanned by 2 waveforms (#1 and #2) containing the Fisher discriminant trained from the first 50 principal components. Illustrated data were pooled from all 18 mice used in this study, which contained 9 wild type and 9 Sp4 hypomorphic animals. Circular symbols and ellipses represent the grand averages and 1 boundaries of the covariances for each genotype groups. Green line marks the decision boundary for classification of the genotype with the least prediction error. (c) The exact same projection plot as (b), except that individual trials are replaced by the mean (circles and squares, representing female and male animals respectively) and covariances (ellipses as 1 σ contours) of individual animals. (d) Cross-validation was performed to determine the optimal number of features (PCs) to use for classification of genotype. For each number of trials to average (vertical axis), the optimum number of features (white line) giving the least prediction error was used for building the linear classifier. (e) Performance of genotype classification based on ERP features is plotted against the number of trials to average. Three methods are illustrated: peaks of a single channel (blue), peaks of multiple channels (red) and PCs of multiple channels (black). Data from 5 channels, FL, FR, PR, OL and OR were used for the multi-channel cases, and channel FR was used for the single channel case.