Figure 3.
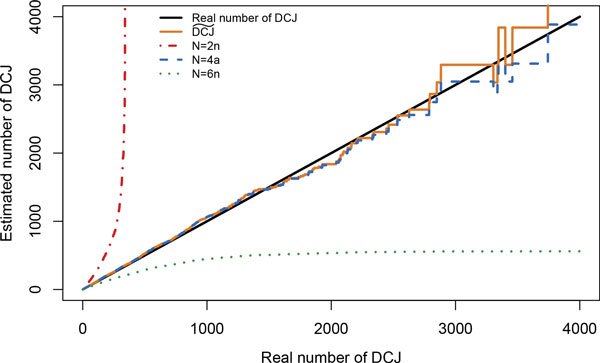
4 × 4 correction when estimating a distance through binary sequence evolution. A genome with a = 980 observed adjacencies and n = 1020 adjacencies in total was evolved with DCJ. The set of observed adjacencies was coded in a sequence, with a 1 for the presence of an adjacency and 0 for the absence. The number of observable sites, that is, the number of 0 s, was set to N , and variations on N were performed. Choosing a sequence of length N < 4a, where a is the number of observed adjacencies of the genomes, leads to an overestimation, while N > 4a leads to an underestimation. With N = 4a, as the theoretical results predicted, the estimation is correct and its quality is equivalent to the direct prediction.
