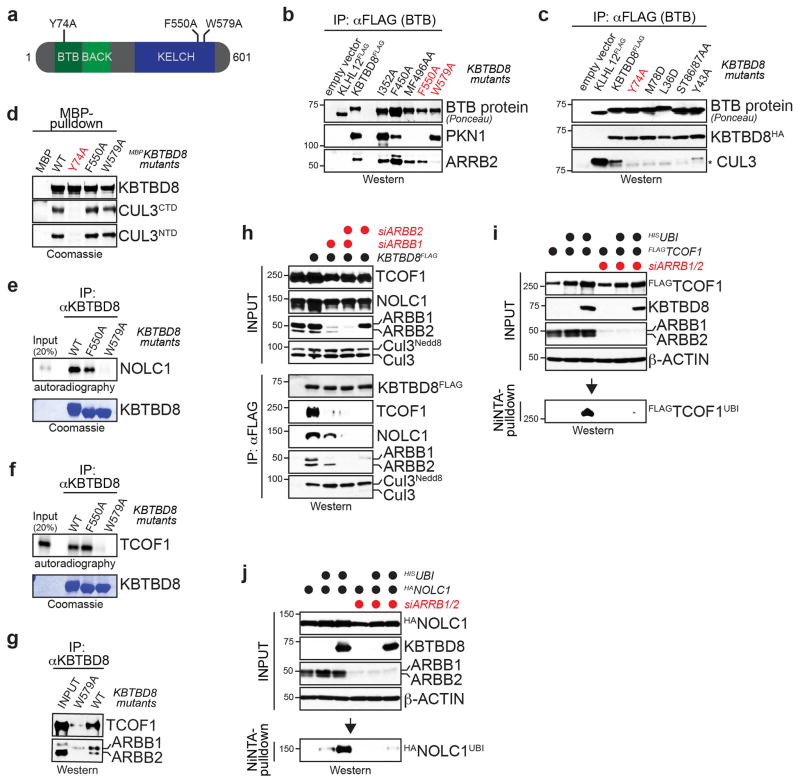
Extended Figure 5. Biochemical characterization of the substrate adaptor role of KBTBD8.
a. Domain structure of KBTBD8, including the residues mutated to generate ubiquitylation- (Y74A) and substrate-binding-deficient KBTBD8 (F550A; W579A). b. Effects of point mutations in predicted KELCH domain loops on binding of KBTBD8 to candidate substrates were determined by affinity-purification and Western. c. Effects of point mutations in BTB domain on binding of KBTBD8 to CUL3 were determined by affinity-purification and Western. Dimerization of FLAGKBTBD8 with KBTBD8HA was analyzed in the same experiment to provide a folding control. d. Binding of recombinant CUL3 to immobilized recombinant MBPKBTBD8 variants was analyzed by Coomassie. e. Binding of in vitro-transcribed/translated 36S-NOLC1 to immobilized recombinant KBTBD8 variants was analyzed by autoradiography. f. Binding of in vitro-transcribed/translated 36S-TCOF1 to immobilized recombinant KBTBD8 variants was analyzed by autoradiography. g. Endogenous β-arrestin proteins in reticulocyte lysates binds immobilized, recombinant KBTBD8, as detected by Western. h. 293T cells were transfected with control- or β-arrestin 1/2-siRNAs and reconstituted with FLAGKBTBD8. Binding of KBTBD8 to endogenous TCOF1 and NOLC1 was analyzed by αFLAG-affinity purification and Western. i. Ubiquitylation of HATCOF1 in 293T cells depleted of β-arrestin 1/2 and reconstituted with KBTBD8 was determined after denaturing NiNTA-purification by Western blotting as described above. i. Ubiquitylation of HANOLC1 was detected in 293T cells depleted of β-arrestins and reconstituted with KBTBD8, as described above.