Abstract
A number of studies have been carried out to investigate the association of X-ray repair complementing defective repair in Chinese hamster cells 6 (XRCC6) polymorphisms and cancer risks, and the results remained inconsistent and inconclusive.
To assess the effect of XRCC6 polymorphisms on cancer susceptibility, we conducted a meta-analysis, up to May 23rd 2014, 6267 cases with different types of tumor and 7536 controls from 20 published case–control studies. Summary odds ratios and corresponding 95% confidence intervals for XRCC6 polymorphism and cancer risk were estimated using fixed- or random-effects models when appropriate. Heterogeneity was assessed by chi-squared-based Q-statistic test, and the sources of heterogeneity were explored by subgroup analyses, logistic meta-regression analyses and Galbraith plot. Publication bias was evaluated by Begg funnel plot and Egger test. Sensitivity analyses were also performed.
The rs2267437 polymorphism was associated with a significant increase in risks of overall cancers, breast cancer, renal cell carcinoma and hepatocellular carcinoma, and it could increase the cancer risk in Asian population; the rs5751129 polymorphism could increase the cancer risk in overall cancers; the rs132770 polymorphism was associated with the increased renal cell carcinoma risk; furthermore, the rs132793 polymorphism could decrease breast cancer risk and increase risks in “other cancers”.
Overall, the results provided evidences that the single nucleotide polymorphisms in XRCC6 promoter region might play different roles in various cancers, indicating different cancers have different tumorigenesis mechanisms. Our studies may perhaps supplement for the disease monitoring of cancers in the future, and additional studies to determine the exact molecular mechanism might provide us with interventions to protect the susceptible subgroups.
INTRODUCTION
Cancer is a major public health problem in the world, and it is among the leading causes of death over world. In the year of 2014, there will be 1,665,540 new cancer cases and 585,720 cancer deaths expected to occur; of these, 51.3% of the cases and 52.9% of the deaths occurred in males.1 Cancer survival tends to be poor, and most cancers are diagnosed at a late stage and the limited efficient treatments due to the unclear cancer pathogenesis mechanisms.
Genomic instability is one cause of carcinogenesis in human cancers, which could promote various mutations. DNA double-strand break (DSB) is the most serious type of damage, and the unpaired or incorrectly repaired DSB could lead to genomic instability.2,3 In mammalian cells, DSB can be repaired mainly by non-homologous end joining (NHEJ) pathway, involving several DSB repair genes, such as X-ray repair complementing defective repair in Chinese hamster cells 6 (XRCC6).4 Genetic variations in NHEJ genes, such as single nucleotide polymorphism (SNP) might escape cell checkpoint surveillance, and can lead to suboptimal DNA repair, allowing DNA damage to accumulate, and finally trigger tumor initiation.5–7
XRCC6 is a gene coding Ku70 protein. XRCC6 gene is one component of NHEJ pathway. It plays an important role in suppression of chromosomal rearrangements and maintenance of genome integrity, thus it is considered essential for genome stability and cell survival. Genetic polymorphism in the XRCC6 gene is hypothesized to have a critical role in tumorigenesis. A number of studies have been carried out to investigate the association of XRCC6 polymorphisms and cancer risks, such as breast cancer,8,9 lung cancer,10 hepatocellular carcinoma (HCC),11,12 glioma,13 and so on, however, the results are inconsistent and inconclusive. Also, several functional studies have been carried out to demonstrate the effects of XRCC6 polymorphisms on Ku70 transcriptional activity in limited types of cancers.14–16 The overall influence of XRCC6 polymorphisms on cancer risks is still ambiguous.
Until recently, the meta-analyzes assessing the association between XRCC6 polymorphisms and cancer17–19 are limited to one single polymorphism site. Systematic review of association between XRCC6 polymorphisms and cancer risk is lacking, and the role of XRCC6 in the etiology of cancer is still equivocal.
We carried out a meta-analysis on all updated published case–control studies to estimate the overall tumor risk of XRCC6 polymorphisms in Asian and European population and to investigate heterogeneity between the individual studies as well as the existence of potential publication bias. The results provided evidences that the SNPs in XRCC6 promoter region might associate with the cancer risks, while SNPs in the XRCC6 intron might not.
MATERIALS AND METHODS
Selection of Published Studies
Studies dealing with the association between cancer risk and XRCC6 gene polymorphisms were considered eligible. We searched the PubMed, Web of Science and Embase databases for all articles on the association between XRCC6 polymorphism and cancer risk (last search update May 23rd 2014). The following terms were used in this search: “Ku70, XRCC6 or X-ray repair complementing defective repair in Chinese hamster cells 6” and “cancer, carcinoma or tumor” and “polymorphism or polymorphisms”. Additional eligible studies on this topic were identified by a hand search of references of retrieved articles. Studies testing the association between XRCC6 gene polymorphism and cancer were included if all the following conditions were met5,8–14,16,20–30: the publication was a case–control study; the study provided the total number of cases and controls; the study provided available genotype frequency in case and control group, respectively; the study was published in English. The major exclusion criteria were as follows: duplicate data31; abstract, comment, review or editorial.17–19 Finally, 20 case–control studies including 6267 cancer cases and 7536 cancer-free controls were included in this meta-analysis, studying the association between different XRCC6 polymorphisms with various cancer risks in Asian and European population. The publication year of eligible studies ranged from 2003 to 2014. Among them, 17 studies concerning XRCC6 C1310G (rs2267437) polymorphism (case/control: 5061/6406), 7 studies concerning T-991C (rs5751129) polymorphism (case/control: 1875/3133), 9 studies concerning A-31G (rs132770) polymorphism (case/control: 4768/3517), 6 studies concerning intron 3 (rs132774) polymorphism (case/control: 1468/2738), 4 studies concerning A46922G (rs132793) polymorphism (case/control: 1824/1807), respectively. Polymorphisms of rs132778, rs12163239, and rs6519265 with only 1 concerned article were excluded.
No contacts with authors were carried out. Ethical approval and informed patient consent was not required as this study was a literature review and had no direct patient contact or influence on patient care.
Data Extraction
Two investigators (Ren and Yan) independently extracted data from each study with a predefined review form, and discrepancies were resolved by consensus of all investigators. All study personnel were blinded throughout the meta-analysis. The following information was recorded for each study: the surname of first author, year of publication, cancer type, ethnicity, number of cases and controls, genotyping methods, matching variables, minor allele frequency in controls, and status of Hardy–Weinberg equilibrium (HWE).
To assess the quality of each eligible case–control study, the same two investigators (Ren and Yan) worked independently to determine the adequacy of studies selection, and discrepancies resolved by discussion with all investigators. All assessors were blinded throughout the meta-analysis. The followings were assessed: the cases and control definition; the comparability of cases and controls on the basis of the design or analysis; the genotyping examinations of polymorphisms; the ascertainment methods for cases and controls; the sources of control; and HWE status of controls.
Cases and controls were marched in all the 20 studies. Among them, 12 studies provided age- gender-marched controls, 5 studies provided additional factor marched controls besides age and gender, such as ethnic background, residence area, individual habits, and 3 studies provided gender-marched controls (Tables 1–5).
TABLE 1.
Characteristics of Literatures on XRCC6 rs2267437 Polymorphism Included in the Meta-Analysis
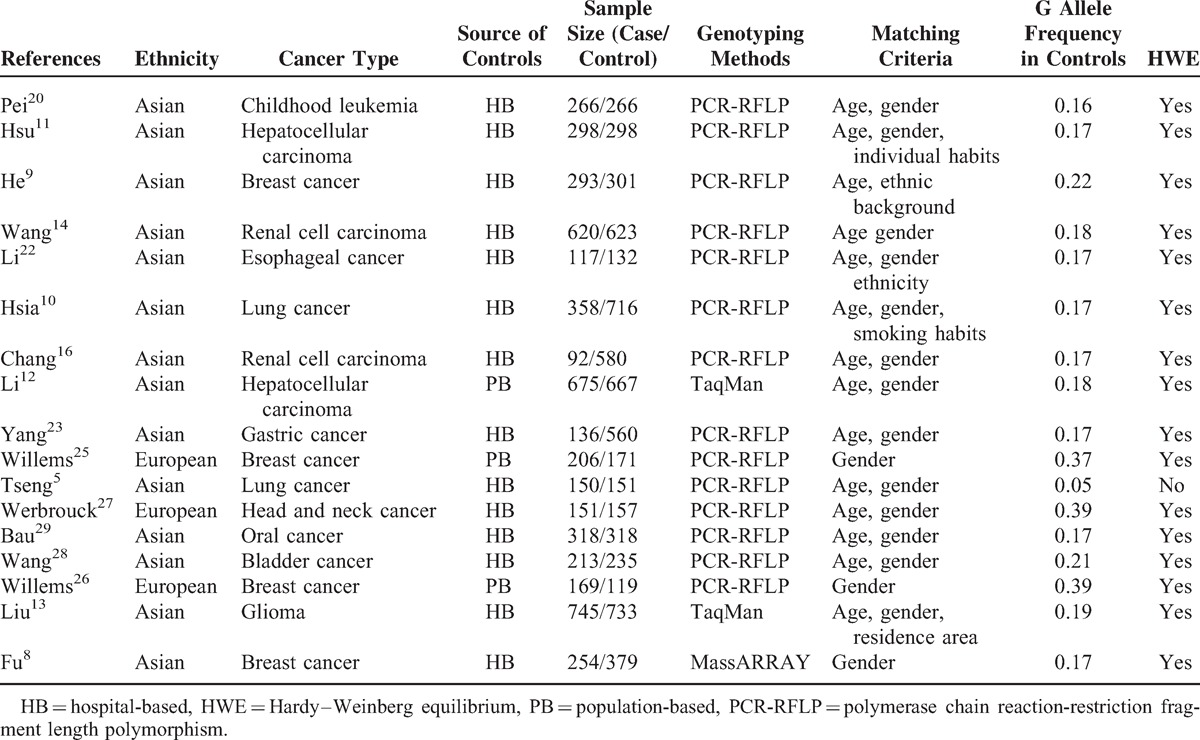
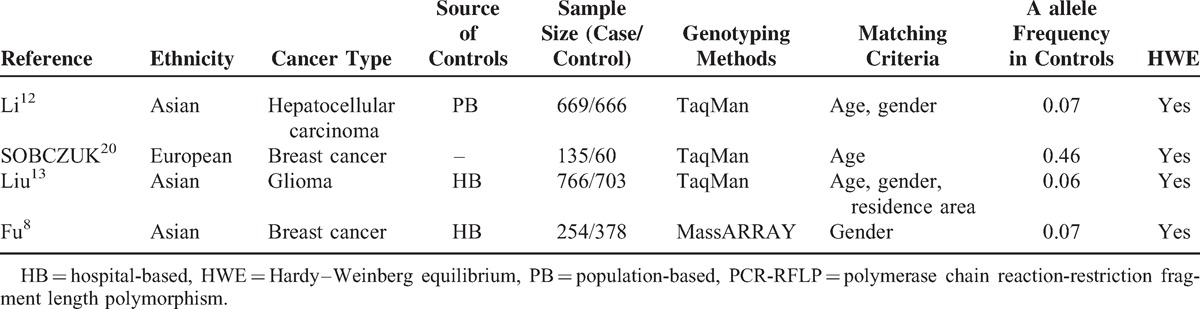
TABLE 5.
Characteristics of Literatures on XRCC6 rs132793 Polymorphism Included in the Meta-Analysis
Statistical Analysis
Deviation from HWE in the control group was examined by χ2 test, P < 0.05 was considered to be statistically significant.32 The strength of relationship association between XRCC6 polymorphisms and cancer risk was assessed by using the pooled odds ratio (OR) with 95% confidence intervals (95% CI). The significance of the pooled OR was determined by Z test, with P < 0.05 considered statistically significant. We evaluated the risk using the allele model (A vs. a), the homozygous model (AA vs. aa), the heterogeneity model (Aa vs. aa), the dominant model (AA + Aa vs. aa), and the recessive model (AA vs. Aa + aa). The chi-squared-based Q-statistic test was used to assess heterogeneity. When the result of the heterogeneity test was P < 0.05, the random-effects model was used (the DerSimonian and Laird method).33 Otherwise, the fixed-effects model was selected (the Mantel–Haenszel method).34 To explore sources of heterogeneity across studies, we did subgroup analyses and logistic meta-regression analyses.35 Galbraith plot was also used to explore sources of heterogeneity when meta-regression analyses could not find the heterogeneity source.36 Subgroup analysis based on ethnicity, cancer types (if one cancer type contains only one study, it was merged into the “other cancers” group), source of controls (hospital-based studies and population-based studies), HWE status of controls (HWE consistent: P > 0.05 and HWE inconsistent: P < 0.05) was performed. Funnel plots and Egger linear regression were used to diagnose a potential publication bias.37 For the possible publication bias, trim and fill method was used to evaluate the influence to the result.38 Sensitivity analyses were performed to assess the stability of the results by excluding one study at a time. All analyses were done, using STATA software, version 10.0 (STATA Corp., College Station, TX, USA). All graphs were obtained also by STATA software. All the P values were two-sided. Data from this meta-analysis are presented in accordance with the checklist proposed by the Meta-Analysis of Observational Studies in Epidemiology group.39
RESULTS
Characteristics of Studies
Overall, 20 studies including 6267 cases and 7536 controls, concerning 5 XRCC6 polymorphisms were included in this meta-analysis. Study characteristics are summarized in Tables 1, 2, 3, 4 and 5.
TABLE 2.
Characteristics of Literatures on XRCC6 rs5751129 Polymorphism Included in the Meta-Analysis
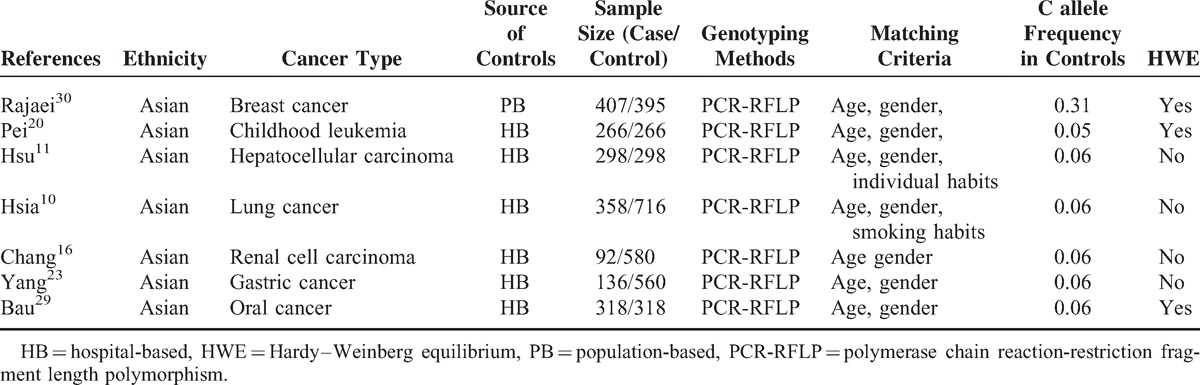
TABLE 3.
Characteristics of Literatures on XRCC6 rs132770 Polymorphism Included in the Meta-Analysis
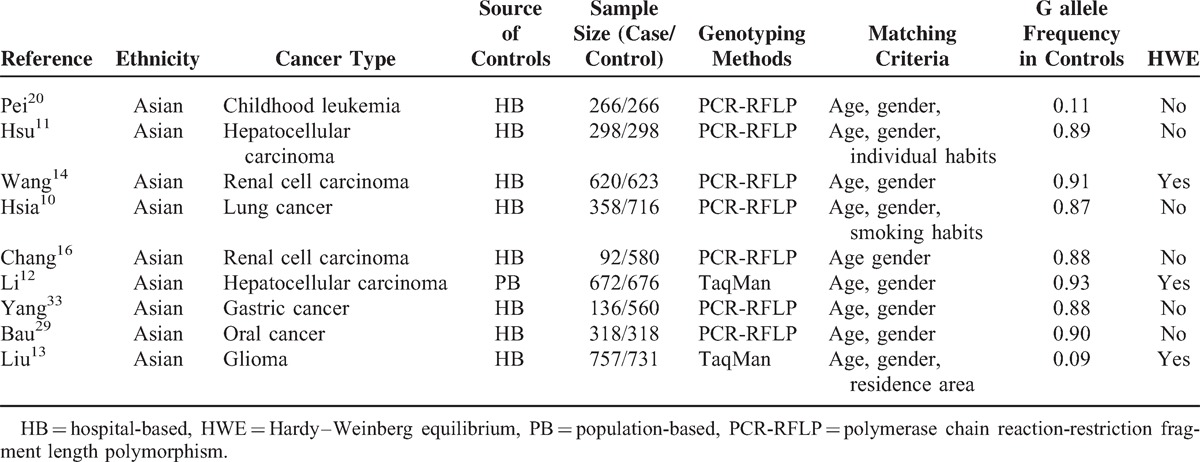
TABLE 4.
Characteristics of Literatures on XRCC6 rs132774 Polymorphism Included in the Meta-Analysis
The 20 articles concerned XRCC6 C1310G (rs2267437) polymorphism, T-991C (rs5751129) polymorphism, A-31G (rs132770) polymorphism, intron 3 (rs132774) polymorphism, A46922G (rs132793) polymorphism, respectively. Among the 20 studies, 6 investigated breast cancer, 3 investigated renal cell carcinoma (RCC), 2 investigated HCC, 2 investigated lung cancer, and 1 for oral cancer, childhood leukemia, esophageal cancer, gastric cancer, bladder cancer, glioma, head and neck cancer each. There were 4 studies of European descendents, 16 studies of Asian descendents, among which 15 studies were of Chinese population. For the selection of controls, 4 studies were population-based case–control studies, while the remaining 16 were hospital-based case–control studies.
Several genotyping methods were used, including Taq-Man, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), MassARRAY. 60% (12/20) of these studies included described genotyping quality control measures, such as positive and negative controls, blindness to the case–control status, a different genotyping assay to confirm the data, and/or random repetition of a portion of samples.
Association Between the XRCC6 Polymorphisms and Cancer Risk
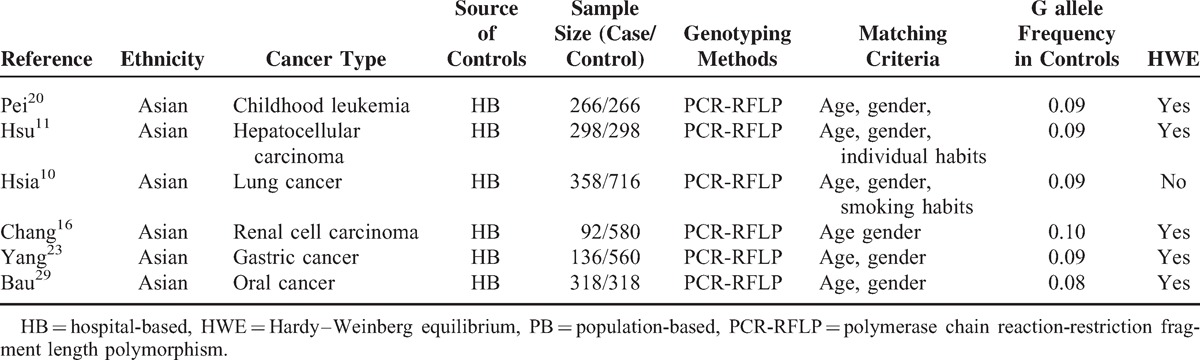
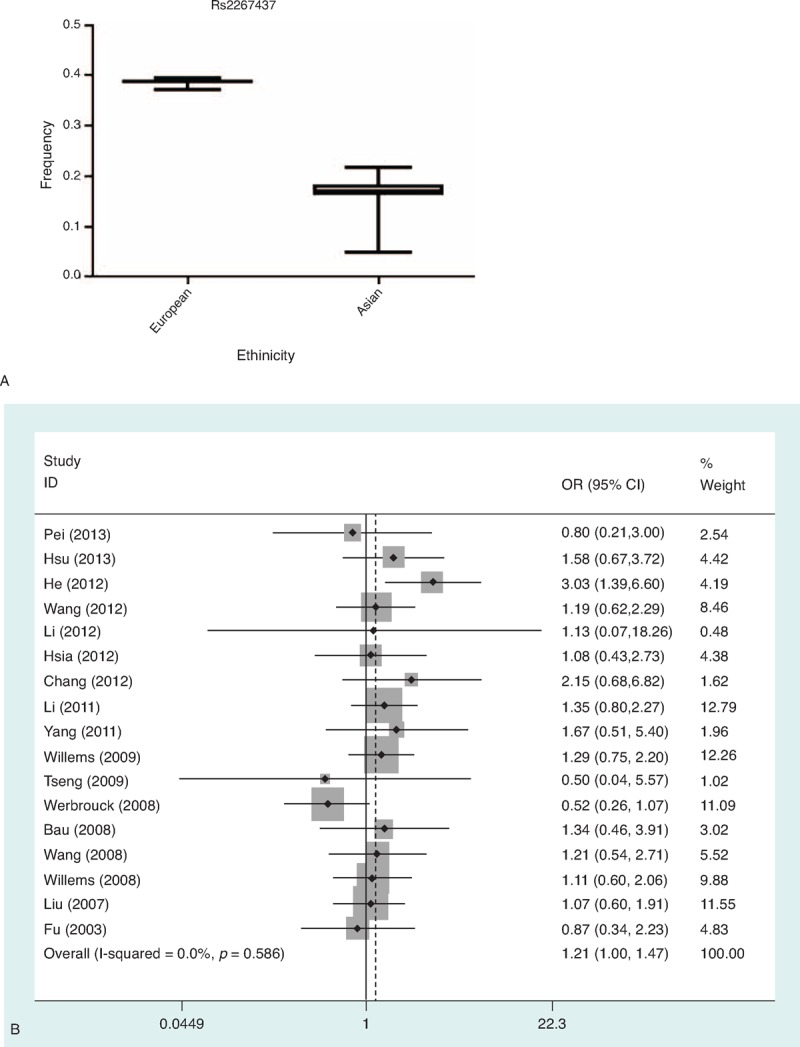
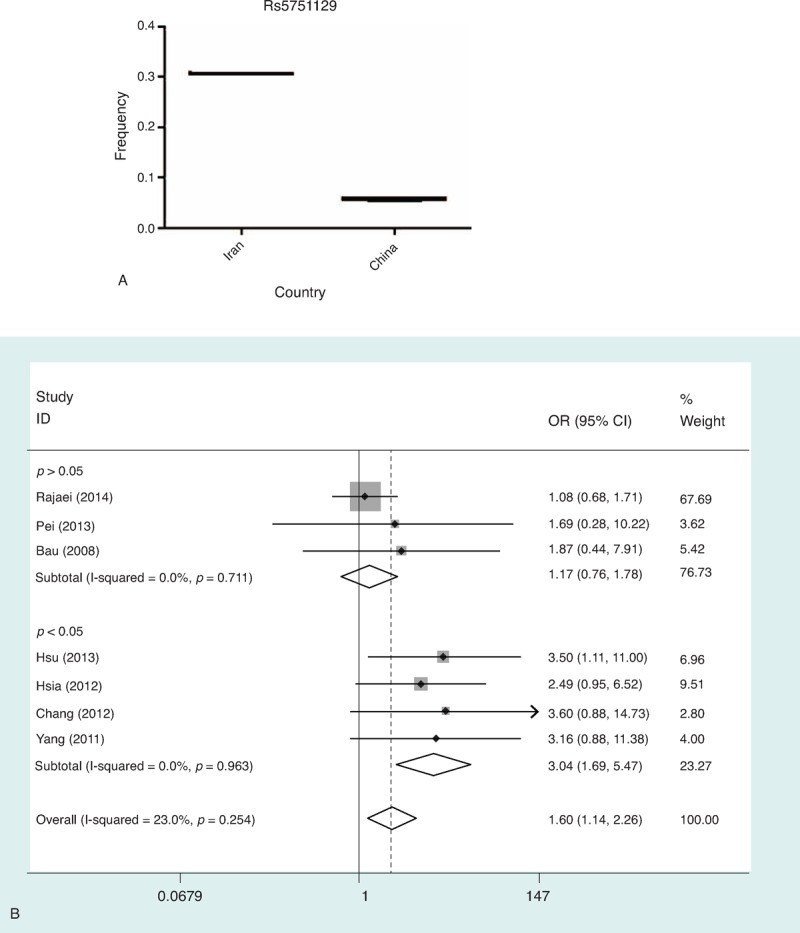
The G allele frequency in rs2267437 polymorphism varied widely between Asian and European ethnicities, ranging from 0.05 in an Asian population to 0.39 in a European Population. The mean frequency of G allele was 0.17 for Asian, and 0.38 for European (Figure 1A, Table 1). The C allele or G allele frequency in rs5751129 polymorphism or rs132793 polymorphism also varied widely in different populations (Figures 2A and 4A).
FIGURE 1.
A. Frequencies of the XRCC6 rs2267437 G allele among control subjects stratified by ethnicity. B. Forest plot of cancer risk associated with the GG genotypes compared with the CC/CG genotype in XRCC6 rs2267437 polymorphism.
FIGURE 2.
A. Frequencies of the XRCC6 rs5751129T/C among control subjects stratified by nation. B. Forest plot of cancer risk associated with the CC genotypes compared with the TT genotype in XRCC6 rs5751129 polymorphism in subgroup analysis of HWE status.
FIGURE 4.
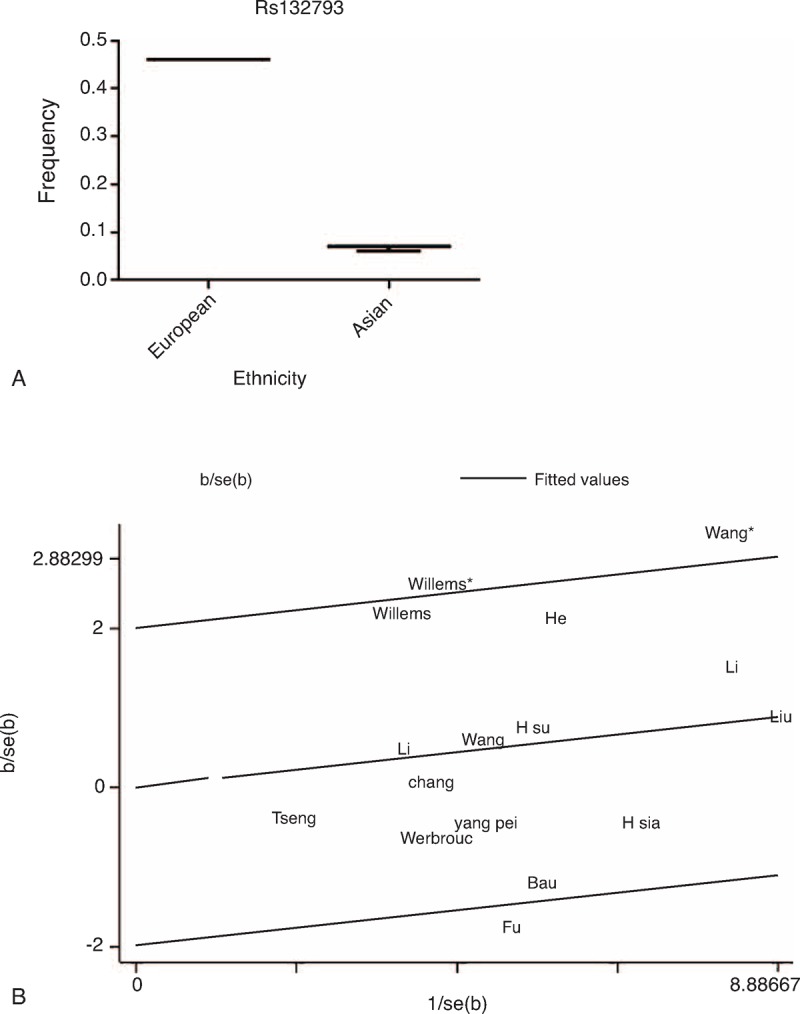
A. Frequencies of the XRCC6 rs132793 among control subjects stratified by country. B Identification of studies acting as sources of heterogeneity by the Galbraith plot under the heterozygous model in rs2267437 polymorphism. Each name represents a separate study for the indicated association. The random effects model was used. Wang∗: 14, Willems∗: 25.
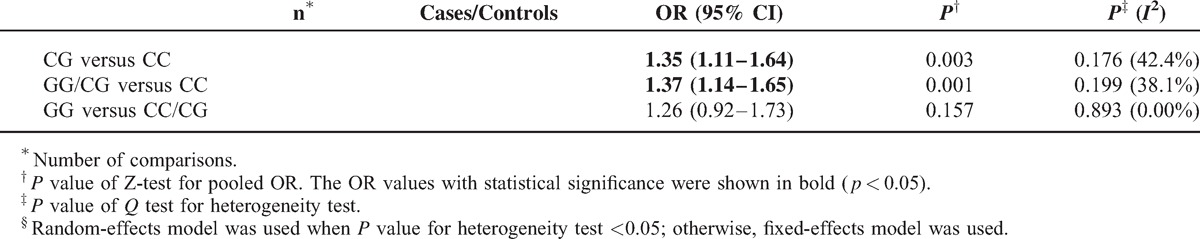
In overall comparison, there was obvious evidence of an association between XRCC6 rs2267437 polymorphism and increased cancer risk (GG vs. CC: OR = 1.33; 95% confidence intervals (CI), 1.09–1.62; recessive model (GG vs. CC/CG): OR = 1.21; 95% CI 1.00–1.47) (Table 6 and Figure 1B). There was also obvious evidence of an association between XRCC6 rs5751129 polymorphism and increased cancer risk (C vs. T: OR = 1.93, 95% CI 1.42–2.63; CC vs. TT: OR = 1.60; 95% CI, 1.14–2.26; TC vs. TT: OR = 1.95, 95% CI 1.48–2.57; dominant model: OR = 2.00, 95% CI 1.50–2.67; recessive model: OR = 1.48, 95% CI 1.06–2.06) (Table 7).
TABLE 6.
Stratified Analyses of the XRCC6 rs2267437 Polymorphism on Cancer Risk
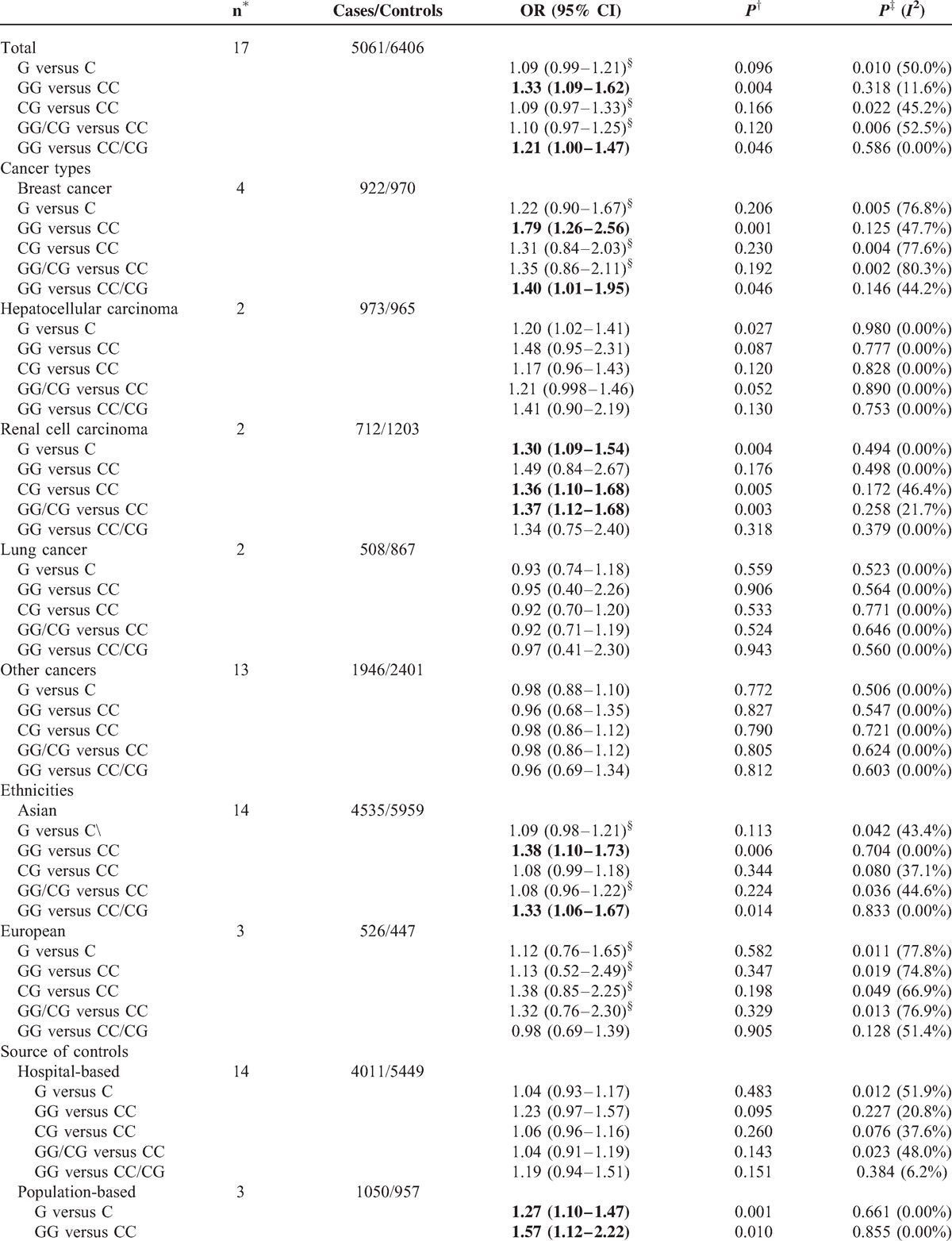
TABLE 6 (Continued).
Stratified Analyses of the XRCC6 rs2267437 Polymorphism on Cancer Risk
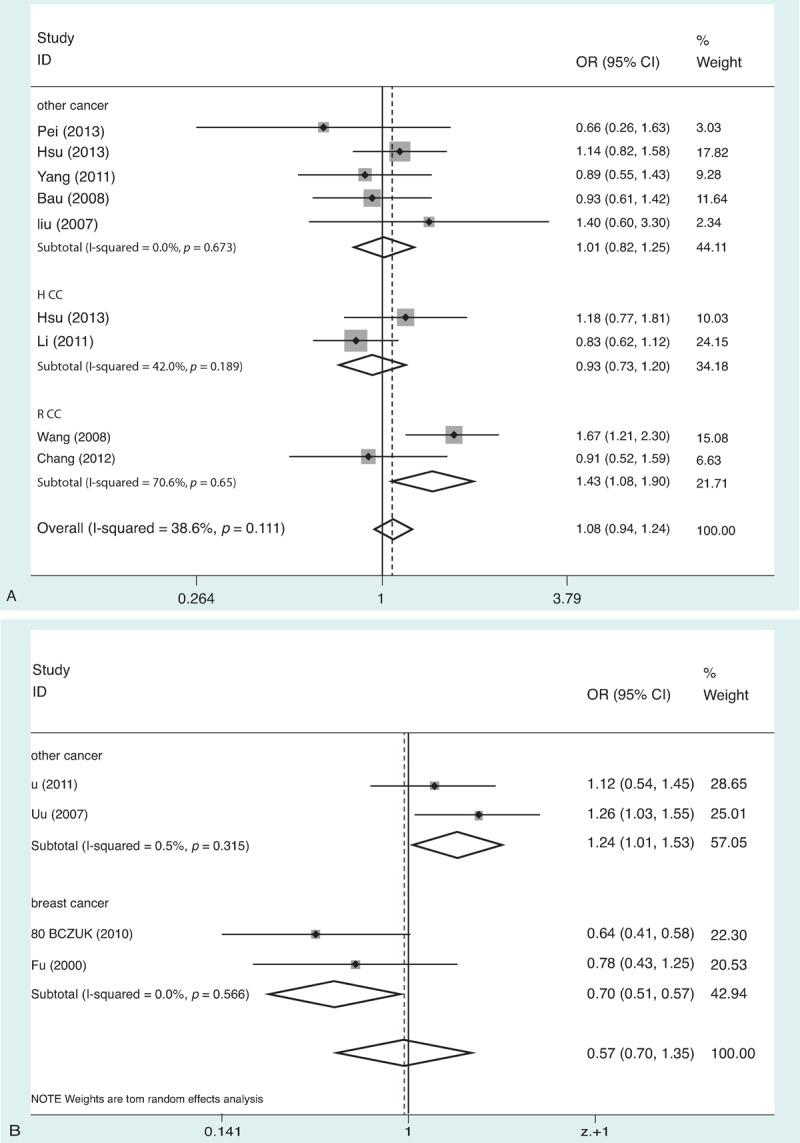
In subgroup analysis, polymorphism rs2267437 was found to be associated with increased cancer risk in breast cancer (GG vs. CC: OR = 1.79, 95% CI 1.26–2.56; recessive model: OR = 1.40, 95% CI 1.01–1.95), HCC (G vs. C: OR = 1.20, 95% CI 1.02–1.41), and RCC (G vs. C: OR = 1.30, 95% CI 1.09–1.54; CG vs. CC: OR = 1.36, 95% CI 1.10–1.68; dominant model: OR = 1.37, 95% CI 1.12–1.68). Similarly, significantly increased risks were observed in the Asian population (GG vs. CC: OR = 1.38, 95% CI 1.10–1.73; recessive model: OR = 1.33, 95% CI 1.06–1.67) in rs2267437 polymorphism (Table 7). The rs132770 polymorphism might associate with increased RCC risk (G vs. A: OR = 1.40, 95% CI 1.08–1.80; recessive model: OR = 1.44, 95% CI 1.08–1.90) (Table 8, Figure 3A). Interestingly, significantly decreased risks were observed in breast cancer in rs132793 polymorphism (A vs. G: OR = 0.70, 95% CI 0.51–0.97), however increased risks were found in “other cancers” in the same polymorphism (A versus G: OR = 1.24, 95% CI 1.01–1.53; GA vs. GG: OR = 1.29, 95% CI 1.03–1.60; dominant model: OR = 1.28, 95% CI 1.03–1.58) (Table 10 and Figure 3B).
TABLE 7.
Stratified Analyses of the XRCC6 rs5751129 Polymorphism on Cancer Risk
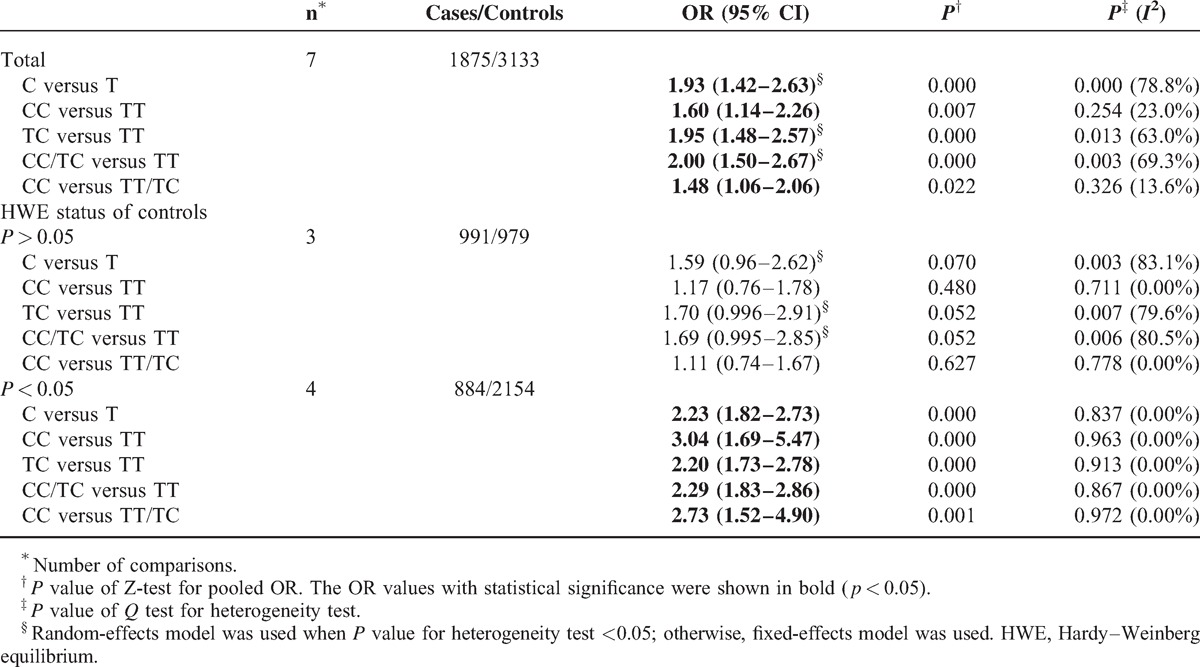
FIGURE 3.
A. Forest plot of cancer risk associated XRCC6 rs132770 polymorphism in subgroup analysis of cancer type in recessive model. B. Forest plot of cancer risk associated with the A genotypes compared with the G genotype in XRCC6 rs132793 polymorphism in subgroup analysis of cancer type.
TABLE 9.
Stratified Analyses of the XRCC6 rs132774 Polymorphism on Cancer Risk
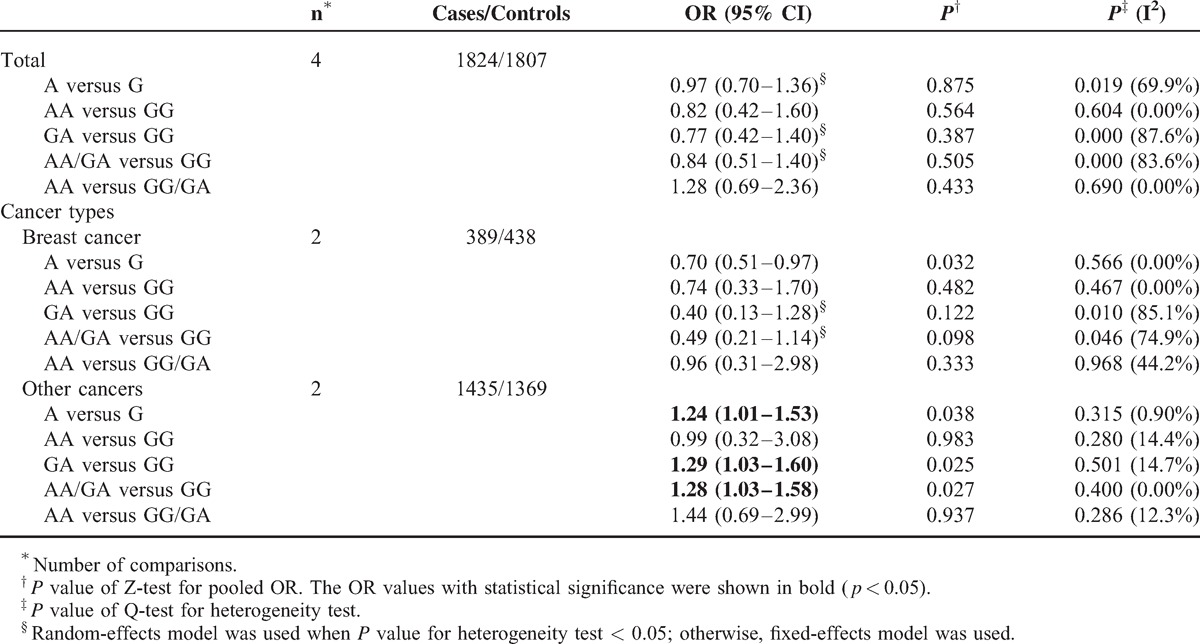
In addition, when stratified by HWE status of controls. Significantly elevated risks were observed in HWE inconsistent studies of rs5751129 polymorphism (C vs. T: OR = 2.23, 95% CI 1.82–2.73; CC vs. TT: OR = 3.04; 95% CI, 1.69–5.47; TC vs. TT: OR = 2.20, 95% CI 1.73–2.78; dominant model: OR = 2.29, 95% CI 1.83–2.86; recessive model: OR = 2.73, 95% CI 1.52–4.90) (Table 7, Figure 2B). No associations were found in HWE consistent studies.
After stratified separately by “sources of control”, significantly elevated risk was found in population-based studies in all comparison models tested except for recessive model of rs2267437 polymorphism (G vs. C: OR = 1.27, 95% CI 1.10–1.47; GG vs. CC: OR = 1.57, 95% CI 1.12–2.22; CG vs. CC: OR = 1.35, 95% CI 1.11–1.64; dominant model: OR = 1.37 95% CI 1.14–1.65) (Table 7).
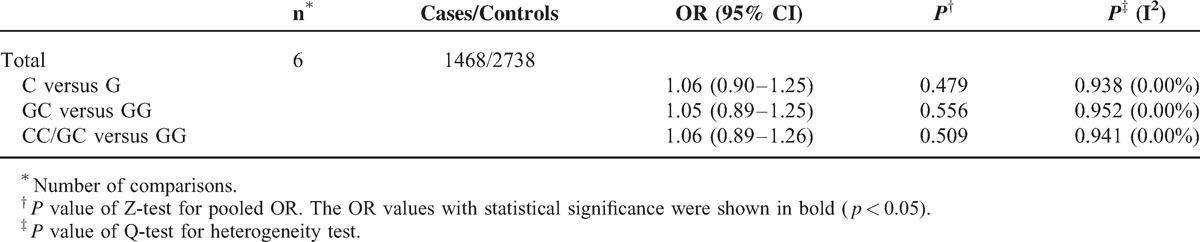
No association was found between the XRCC6 rs132774 polymorphism and cancer risk (Table 9).
TABLE 8.
Stratified Analyses of the XRCC6 rs132770 Polymorphism on Cancer Risk
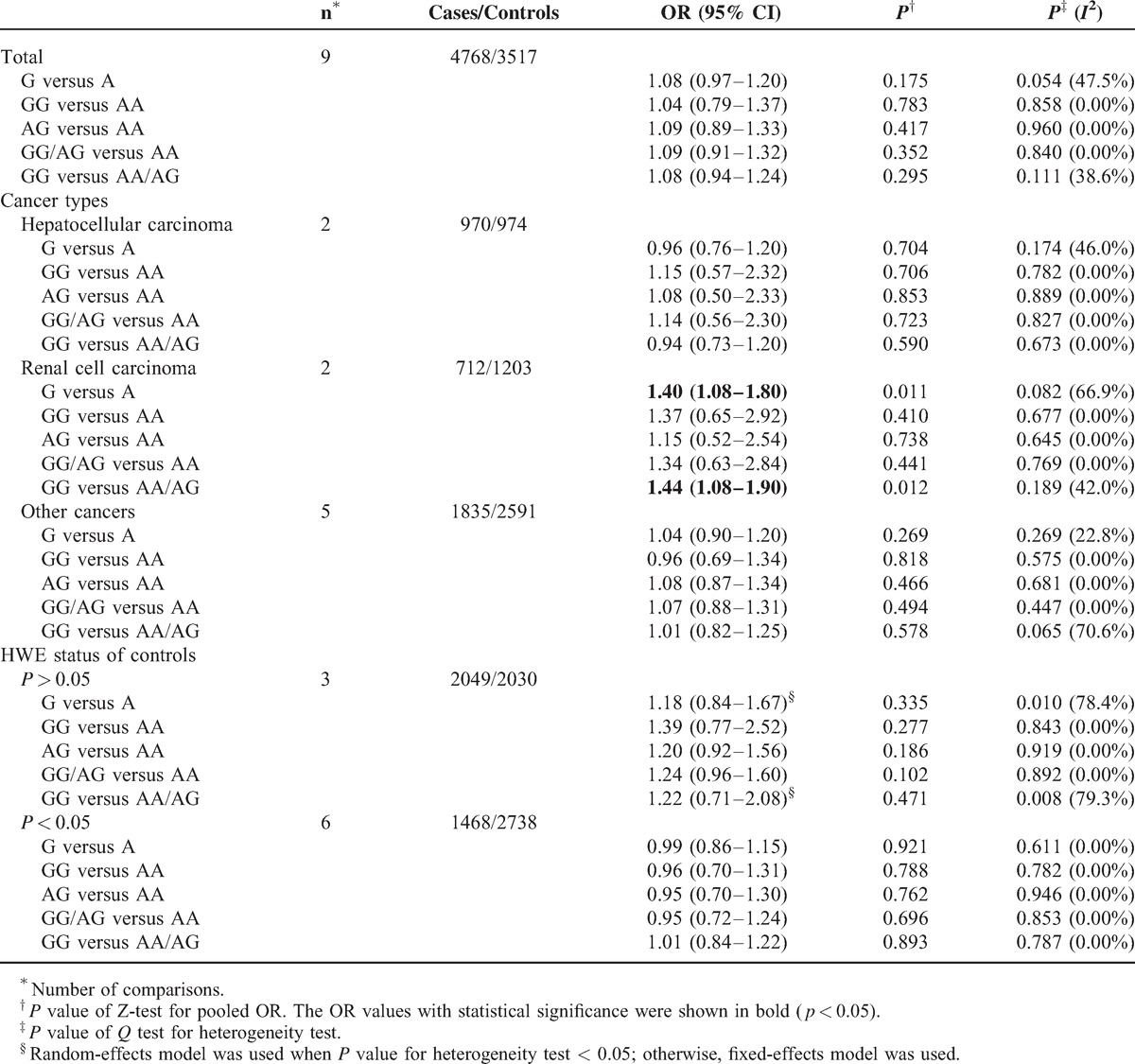
TABLE 10.
Stratified Analyses of the XRCC6 rs132793 Polymorphism on Cancer Risk
Evaluation of Heterogeneity
There was heterogeneity among studies in overall comparisons and also subgroup analyses in 3 XRCC6 polymorphisms: rs2267437, rs5751129, and rs132793. To explore sources of heterogeneity, we evaluated the following variables: ethnicities, cancer type, source of control, study quality, genotyping methods and sample size (≤500 and >500 subjects). Galbraith plot was also used to detect the possible sources of heterogeneity when none of the above variables could explain the heterogeneity.
For the rs2267437 polymorphism, there was significant heterogeneity in overall comparisons of allele model, heterozygote model, and dominant model. None of the possible variables could explain the heterogeneity. The studies potentially causing between-study heterogeneity were identified in the allele model8,9,27), heterozygote model8,14,25, and dominant model8,9,14,25 by the Galbraith plot (Figure 4B). However, the result was altered in allele model (OR = 1.13 95% CI 1.05–1.21) when the studies were excluded.
Meta-regression analysis indicated that the “source of control” could explain 100% of the τ2 in all comparison models in rs5751129 polymorphism, and the “cancer type” could explain 100% of the τ2 in allele model in rs132793 polymorphism. Furthermore, the combination of “ethnicity” and “cancer type” could explain 100% of the τ2 in heterozygote and dominant model in rs132793 polymorphism.
Sensitivity Analysis
Sensitivity analyses were performed by sequential removal of each eligible study to assess the influence of each individual study on the pooled OR in each comparison in the polymorphisms of rs2267437, rs5751129, and rs132793. The omission of any study made no significant difference, indicating that the results of this meta-analysis were statistically reliable (Figure 5A).
FIGURE 5.
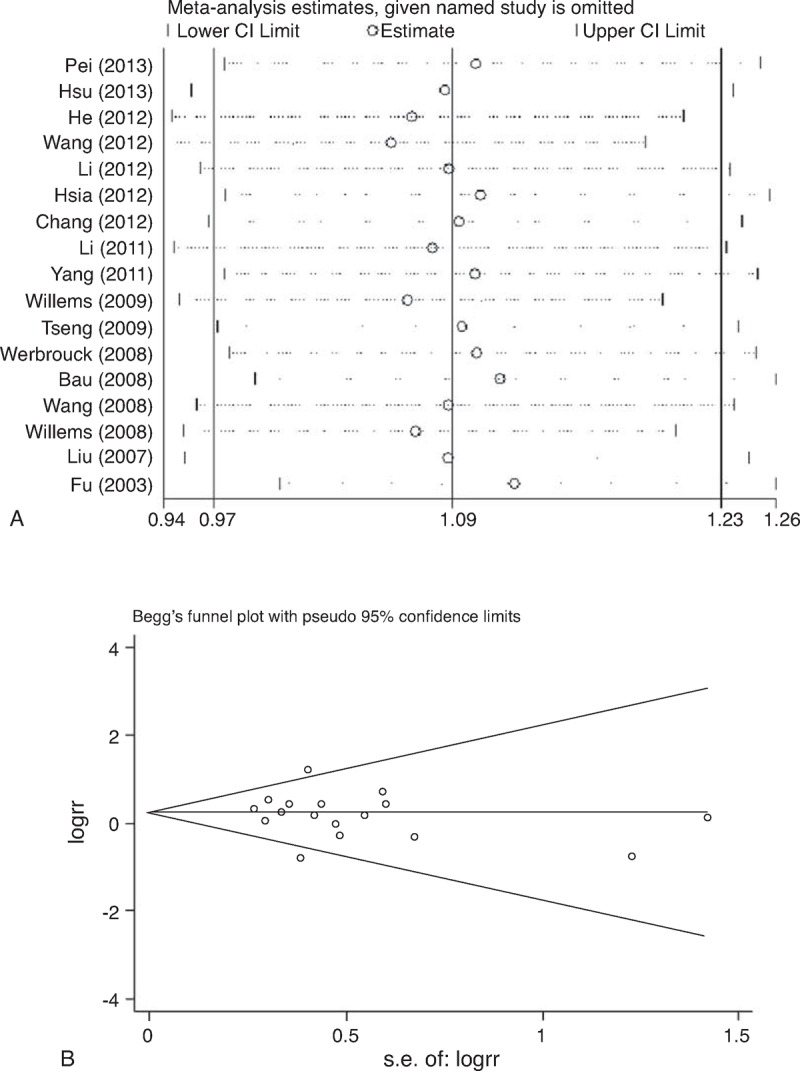
A. Sensitivity analysis of the heterozygous model in XRCC6 rs2267437 polymorphism. B. Begg funnel plot for publication bias test under homozygous model in rs2267437 polymorphism. Each point represents a separate study for the indicated association.
Publication Bias
Egger test reveals evidence of publication bias in rs5751129 polymorphism (Figure 5B). The trim and fill method showed that the funnel plot needs 4 more studies to be symmetrical (Figure 6) in allele model, recessive model and homozygous model, or 2 more in heterozygous and dominant model. But the results were altered in recessive model and homozygous model. The publication bias in rs132793 polymorphism was not checked because of the limited number of relevant studies.
FIGURE 6.
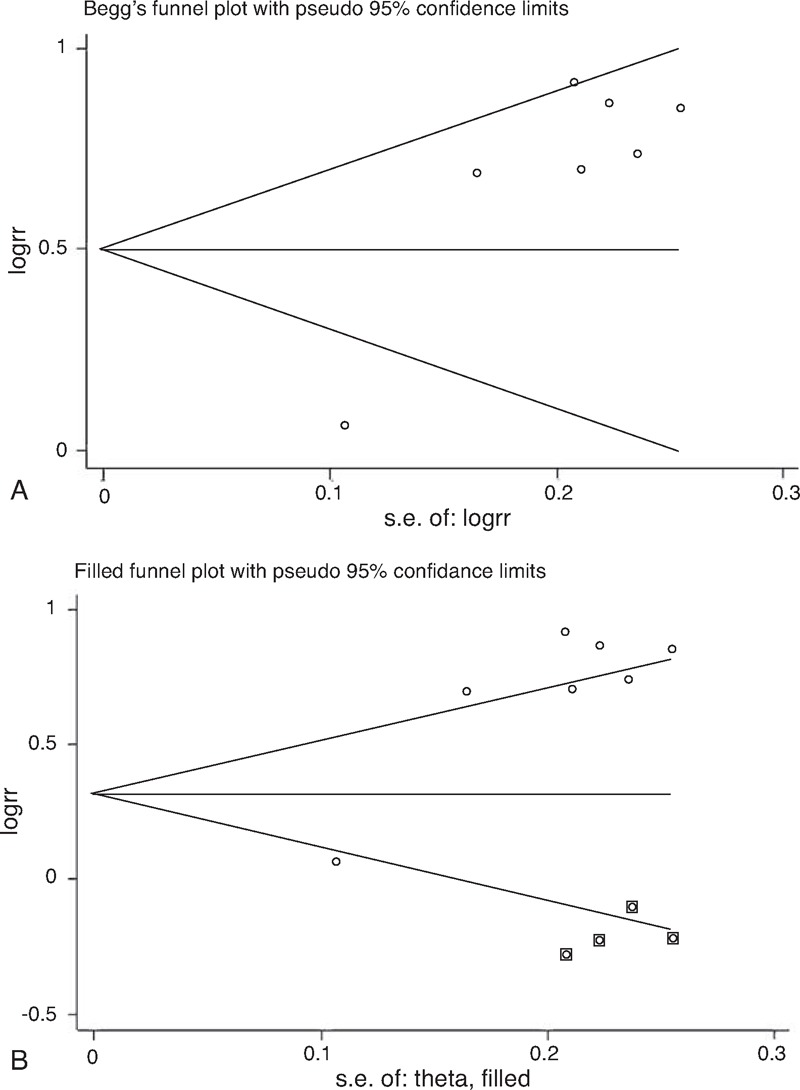
Begg Funnel Plots of Meta-Analysis before (A) and after (B) Trim and Fill Adjustment for Publication Bias under allele model in XRCC6 rs5751129 polymorphism.
DISCUSSION
In this study, we performed a systematic review of association between XRCC6 polymorphisms and cancer risks based on 20 case–control studies. This is also the first time to explore the individual association between the polymorphisms of rs5751129, rs132774, rs132793, and cancer risk. The results provided evidences that the SNPs in XRCC6 promoter region might associate with the cancer risks, while SNP in the XRCC6 intron might not. In addition, the XRCC6 promoter SNPs might play different roles in various cancers, indicating that XRCC6 gene may have different roles in different cancer development, and different mechanisms promote the development of various tumors.
XRCC6 gene was involved in multiple cellular pathways, including NHEJ pathway of DSB repair.40,41 During DNA repair procedure, Ku70 acted as a scaffold to recruit the other NHEJ factors to the damage site after its bound to DNA ends.4 However, inaccurate repair could lead to cellular aberrant function, apoptosis, and chromosomal rearrangements, which promote carcinogenesis finally.42 Besides the influence of XRCC6 gene on DSB repair and genomic stability, it has some other NHEJ-independent effects. Such as the Ku70 protein in cytoplasm can prevent Bax translocate to mitochondria, and suppresses cell apoptosis.43 The defects in Ku70 may also influence cell proliferation.44
It has been demonstrated that the rs2267437 polymorphism could influence the expression level and stability of the Ku70 protein in breast cancer cells and RCC tissues.14,15 The sequence variation in the rs2267437 may affect binding activity of the adjacent CACCC box with transcription factors, resulting in decreased Ku70 expression level, and the DSB repair activity thus was affected, finally leading to increased susceptibility to cancers.45 In our study, rs2267437 polymorphism was found to might increase the cancer risks in breast cancer, HCC and RCC, while may not function in lung cancer or some other cancers, and different ethnicities might influence the association.
The rs5751129 polymorphism locates between rs132770 and rs2267437 in the promoter region of XRCC6 gene. This polymorphism was proved to be functional in HCC and RCC, with the normal tissues with C allele having a lower expression level of XRCC6 mRNA or protein.11,16 Our results indicated that rs5751129 polymorphism might play the same role in various cancers. The variation may influence the expression level or stability of XRCC6 mRNA via alternative spicing or other mechanisms. Individuals carrying C allele may exhibit heritable decreased DNA repair capacity phenotypes compared with those carrying T allele, thus they might have less protective effects on normal tissues, and increased cancer risks. However, when stratified by HWE status of controls, we found that significantly elevated risks were observed in HWE inconsistent studies, but not in HWE consistent studies. Thus there may be possibility that the presence of the C allele is in linkage disequilibrium with another mutation located outside the coding region in the XRCC6 gene, which may be important for the Ku70 expression.
The rs132770 polymorphism locates closer than rs5751129 polymorphism to the translation starting point in the XRCC6 promoter. Our meta-analysis indicating that rs132770 polymorphism might affect RCC risk in a different way from in other cancers, the RCC pathogenesis mechanism might be distinguished from other cancers when rs132770 polymorphism involved. The results were consistent with the finding that the A allele of the rs132770 polymorphism could increase the expression levels of the Ku70 mRNA in normal tissue of RCC patients.21 Xu17 also collected 3 studies but failed to find any association. The negative results in Xu study might be due to the limited studies.
It is interesting that the rs132793 polymorphism was found to play opposite roles in different cancers: it might decrease breast cancer risk, whereas increase cancer risk in “other cancers”, indicating that the rs132793 may play opposite roles in breast cancer contrast to other cancers. The rs132793 locates in the sequences downstream of stop code of XRCC6 gene, and until recently, the role of rs132793 in tumorigenesis is unclear. We hypothesized that the rs132793 polymorphism might influence breast cancer risk via some mechanisms different from other cancers. Functional studies should be carried out to explore the mechanisms involving rs132793 polymorphism in tumorigenesis in the future. The different ethnicities of the study population might influence the genetic effect of the rs132793 polymorphism on cancer susceptibility; anyhow, it seems not substantial in our study.
Our results implied that rs132774 polymorphism have no association with cancer risk in Chinese population. To validate the results, more population-based studies in other populations should be carried out in the future.
There was heterogeneity among studies in rs2267437, rs5751129, and rs132793 polymorphisms. “Source of control” and “ethnicity”, “cancer type” might explain 100% of the τ2 in the rs5751129 and rs132793 polymorphism, respectively. However, the combination of “ethnicities”, “source of control”, and “sample size” could explain only 17.6% of the τ2 in heterozygote model in rs2267437 polymorphism, implying that there may be other reasons accounting for the heterogeneity in the rs2267437 polymorphism. Publication bias was found in the rs5751129 polymorphism and trim and fill method could reduce the influence of publication bias in all models except recessive and homozygous model, implying that more cautions should be paid when elucidate the role of the rs5751129 polymorphism in cancer susceptibility. Anyhow, sensitivity analysis proved that the results of this meta-analysis were statistically reliable. Therefore, a methodologically preferable design, such as using population-based controls, is crucial to avoid selection bias and heterogeneity.
The limitations of our meta-analysis should also be discussed. First, the non-English articles were excluded in our study, which thus may bias the results of our results. Second, some low-quality studies with deviation from HWE in the control group were included in our meta-analysis. Subgroup analysis were carried out by HWE status of controls in rs5751129 polymorphism and rs132770 polymorphism, and the opposite results were found in rs5751129 polymorphism between HWE consistent/inconsistent groups, implying that low-quality studies might influence the results in rs5751129 polymorphism. Third, the number of included studies was relatively small in some subgroups, thus the specific results should be interpreted with caution.
In conclusion, this meta-analysis showed some evidence of the XRCC6 SNP polymorphisms and cancer risk, supporting the existence of association between XRCC6 polymorphisms and cancer risks in different ethnicities and cancer types. The rs2267437 polymorphism was found to be associated with a significant increase in risks of overall cancers, breast cancer, RCC and HCC, and it might increase the cancer risk in Asian population. The rs5751129 polymorphism might increase the cancer risk in overall cancers, and the rs132770 polymorphism might be associated with the increased cancer risk in RCC. Furthermore, the rs132793 polymorphism could decrease breast cancer risk and increase risks in “other cancers”. However, there was no obvious association between XRCC6 polymorphisms and risks of some “other cancer”. This may be due to the limited studies included or limited sample size in “other cancers”. Therefore, more studies with large sample are required to further confirm the results in the future. Functional studies are also needed to elucidate the roles of XRCC6 promoter polymorphisms in cancer pathogenesis. Moreover, gene–environment and gene–gene interaction analyses, as well as haplotype analysis should be carried out to clarify the role of the XRCC6 genes in cancer. Our studies may perhaps supplement for the disease monitoring of cancers in the future, and additional studies to determine the exact molecular mechanism might provide us with interventions to protect the susceptible subgroups.
Footnotes
Abbreviations: CI = confidence interval, DSB = DNA double-strand break, HB = hospital-based, HCC = hepatocellular carcinoma, HWE = Hardy–Weinberg equilibrium, NHEJ = non-homologous end joining, OR = odds ratio, PB = population-based, PCR-RFLP = polymerase chain reaction-restriction fragment length polymorphism, RCC = renal cell carcinoma, SNP = single nucleotide polymorphism, XRCC6 = X-ray repair complementing defective repair in Chinese hamster cells 6.
This work was supported by Zhejiang Provincial Natural Science Foundation of China (grant No. LQ12H16013), the Health and Family Planning commission of Bureau (XKQ-010-001), and the Science Technology Department (No. 2012F10005) of Zhejiang Province, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript The assistance from Associate Professor Linbo Gao in statistical consultation were highly appreciated by the authors.
The authors have no funding or conflicts of interest to disclose.
REFERENCES
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64:9–29. [DOI] [PubMed] [Google Scholar]
- 2.Evdokimova V, Gandhi M, Rayapureddi J, et al. Formation of carcinogenic chromosomal rearrangements in human thyroid cells after induction of double-strand DNA breaks by restriction endonucleases. Endocr Relat Cancer 2012; 19:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie H, Wise SS, Wise JP., Sr Deficient repair of particulate hexavalent chromium-induced DNA double strand breaks leads to neoplastic transformation. Mutat Res 2008; 649:230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2013; 2:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng RC, Hsieh FJ, Shih CM, et al. Lung cancer susceptibility and prognosis associated with polymorphisms in the nonhomologous end-joining pathway genes: a multiple genotype-phenotype study. Cancer 2009; 115:2939–2948. [DOI] [PubMed] [Google Scholar]
- 6.Dombernowsky SL, Weischer M, Freiberg JJ, et al. Missense polymorphisms in BRCA1 and BRCA2 and risk of breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev 2009; 18:2339–2342. [DOI] [PubMed] [Google Scholar]
- 7.Pucci S, Mazzarelli P, Rabitti C, et al. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene 2001; 20:739–747. [DOI] [PubMed] [Google Scholar]
- 8.Fu YP, Yu JC, Cheng TC, et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res 2003; 63:2440–2446. [PubMed] [Google Scholar]
- 9.He W, Luo S, Huang T, et al. The Ku70-1310C/G promoter polymorphism is associated with breast cancer susceptibility in Chinese Han population. Mol Biol Rep 2012; 39:577–583. [DOI] [PubMed] [Google Scholar]
- 10.Hsia TC, Liu CJ, Chu CC, et al. Association of DNA double-strand break gene XRCC6 genotypes and lung cancer in Taiwan. Anticancer Res 2012; 32:1015–1020. [PubMed] [Google Scholar]
- 11.Hsu CM, Yang MD, Chang WS, et al. The contribution of XRCC6/Ku70 to hepatocellular carcinoma in Taiwan. Anticancer Res 2013; 33:529–535. [PubMed] [Google Scholar]
- 12.Li R, Yang Y, An Y, et al. Genetic polymorphisms in DNA double-strand break repair genes XRCC5, XRCC6 and susceptibility to hepatocellular carcinoma. Carcinogenesis 2011; 32:530–536. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Zhang H, Zhou K, et al. Tagging SNPs in non-homologous end-joining pathway genes and risk of glioma. Carcinogenesis 2007; 28:1906–1913. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Pan X, Huo X, et al. A functional polymorphism C-1310G in the promoter region of Ku70/XRCC6 is associated with risk of renal cell carcinoma. Mol Carcinog 2012; 51suppl 1:E183–190. [DOI] [PubMed] [Google Scholar]
- 15.Ouimet M, Cassart P, Lariviere M, et al. Functional analysis of promoter variants in KU70 and their role in cancer susceptibility. Genes Chromosomes Cancer 2012; 51:1007–1013. [DOI] [PubMed] [Google Scholar]
- 16.Chang WS, Ke HL, Tsai CW, et al. The role of XRCC6 T-991C functional polymorphism in renal cell carcinoma. Anticancer Res 2012; 32:3855–3860. [PubMed] [Google Scholar]
- 17.Xu H, Zou P, Chen P, et al. Association between the XRCC6 Promoter rs2267437 polymorphism and cancer risk: evidence based on the current literature. Genet Test Mol Biomarkers 2013; 17:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L, Ju XB, Li P, et al. Association between the Ku70-1310C/G promoter polymorphism and cancer risk: a meta-analysis. Asian Pac J Cancer Prev 2012; 13:683–687. [DOI] [PubMed] [Google Scholar]
- 19.Jiang H, Lin Y, Yang CQ, et al. Quantitative assessment of the association between XRCC6 C1310G polymorphism and cancer risk. Tumour Biol 2013; 34:779–785. [DOI] [PubMed] [Google Scholar]
- 20.Pei JS, Lee YM, Lo HH, et al. Association of X-ray repair cross-complementing-6 genotypes with childhood leukemia. Anticancer Res 2013; 33:5395–5399. [PubMed] [Google Scholar]
- 21.Wang W, Gao Y, Yan F, et al. Association of Ku70 A-31G polymorphism and risk of renal cell carcinoma in a Chinese population. DNA Cell Biol 2012; 31:1314–1320. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Suo Q, He D, et al. Esophageal cancer risk is associated with polymorphisms of DNA repair genes MSH2 and WRN in Chinese population. J Thorac Oncol 2012; 7:448–452. [DOI] [PubMed] [Google Scholar]
- 23.Yang MD, Wang HC, Chang WS, et al. Genetic polymorphisms of DNA double strand break gene Ku70 and gastric cancer in Taiwan. BMC Cancer 2011; 11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobczuk A, Smolarz B, Romanowicz H, et al. Analysis of the polymorphisms in non-homologous DNA end joining (NHEJ) gene Ku70 and Ligase IV in sporadic breast cancer in women. Pol J Pathol 2010; 61:27–31. [PubMed] [Google Scholar]
- 25.Willems P, De Ruyck K, Van den Broecke R, et al. A polymorphism in the promoter region of Ku70/XRCC6, associated with breast cancer risk and oestrogen exposure. J Cancer Res Clin Oncol 2009; 135:1159–1168. [DOI] [PubMed] [Google Scholar]
- 26.Willems P, Claes K, Baeyens A, et al. Polymorphisms in nonhomologous end-joining genes associated with breast cancer risk and chromosomal radiosensitivity. Genes Chromosomes Cancer 2008; 47:137–148. [DOI] [PubMed] [Google Scholar]
- 27.Werbrouck J, De Ruyck K, Duprez F, et al. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat Res 2008; 656:74–81. [DOI] [PubMed] [Google Scholar]
- 28.Wang SY, Peng L, Li CP, et al. Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int J Urol 2008; 15:534–539. [DOI] [PubMed] [Google Scholar]
- 29.Bau DT, Tseng HC, Wang CH, et al. Oral cancer and genetic polymorphism of DNA double strand break gene Ku70 in Taiwan. Oral Oncol 2008; 44:1047–1051. [DOI] [PubMed] [Google Scholar]
- 30.Rajaei M, Saadat I, Omidvari S, Saadat M. Association between polymorphisms at promoters of XRCC5 and XRCC6 genes and risk of breast cancer. Med Oncol 2014; 31:885. [DOI] [PubMed] [Google Scholar]
- 31.Wen TC, Chang TH, Hsing WC, et al. Association of oral cancer susceptibility and DNA double strand break gene Ku70 single nucleotide polymorphisms. Oral Oncol 2009; suppl 3:1. [Google Scholar]
- 32.Grover VK, Cole DE, Hamilton DC. Attributing Hardy–Weinberg disequilibrium to population stratification and genetic association in case-control studies. Ann Hum Genet 2010; 74:77–87. [DOI] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 34.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22:719–748. [PubMed] [Google Scholar]
- 35.Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med 2002; 21:1559–1573. [DOI] [PubMed] [Google Scholar]
- 36.Galbraith RF. A note on graphical presentation of estimated odds ratios from several clinical trials. Stat Med 1988; 7:889–894. [DOI] [PubMed] [Google Scholar]
- 37.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000; 56:455–463. [DOI] [PubMed] [Google Scholar]
- 39.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000; 283:2008–2012. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Xie M, Li R, et al. Role of Ku70 in deubiquitination of Mcl-1 and suppression of apoptosis. Cell Death Differ 2014; 21:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puebla-Osorio N, Kim J, Ojeda S, et al. A novel Ku70 function in colorectal homeostasis separate from nonhomologous end joining. Oncogene 2014; 33:2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beggs AD, Domingo E, McGregor M, et al. Loss of expression of the double strand break repair protein ATM is associated with worse prognosis in colorectal cancer and loss of Ku70 expression is associated with CIN. Oncotarget 2012; 3:1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Naegele JR, Lin SL. The DNA-PK catalytic subunit regulates Bax-mediated excitotoxic cell death by Ku70 phosphorylation. Brain Res 2009; 1296:164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Y, Seidl KJ, Rathbun GA, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 1997; 7:653–665. [DOI] [PubMed] [Google Scholar]
- 45.Hosoi Y, Watanabe T, Nakagawa K, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol 2004; 25:461–468. [PubMed] [Google Scholar]


