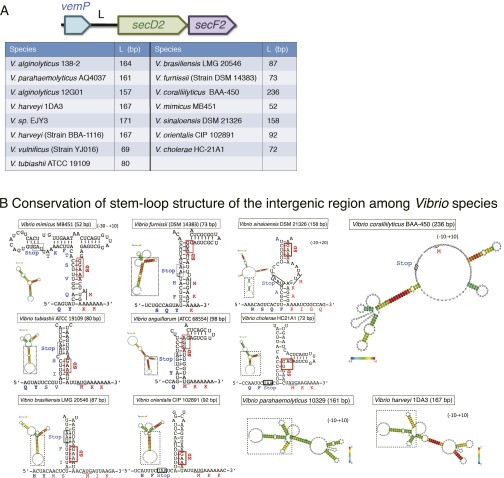
Fig. S6.
Conservation of the stem-loop structures of vemP-secD2 intergenic regions. (A) The vemP-secD2 intergenic regions of different Vibrio strains. Lengths of the intergenic regions between vemP and secD2 from Vibrio species are listed in the table. (B) Predicted secondary structures formed in the intergenic regions. RNA sequences from 10–30 bases upstream of the VemP stop codon to 10–20 bases downstream of the V.SecD2 start codon of various Vibrio species were analyzed by the CentroidHomfold program. The boxed regions of the modeled RNA structures are enlarged to show the stem structures, in which the putative SD sequences of secD2 are highlighted by red boxes. The initiation codons of secD2 are underlined. In the case of V. parahaemolyticus and Vibrio harveyi, only the whole structures are presented, because their core stem structures are almost identical to that from V. alginolyticus (Fig. 6A). Vibrio coralliilyticus BAA-450 is exceptional in having no stem structure in front of the secD2 start codon. In the other strains, we observed several common features in the structures: (i) The start points of the stem structure are very close to the arrest points (a Gln codon near the stop) determined in this study, and (ii) the putative SD sequences of V.SecD2 are engaging in the stem structures. These features are in good agreement with our model of regulation of V.SecDF2 translation.