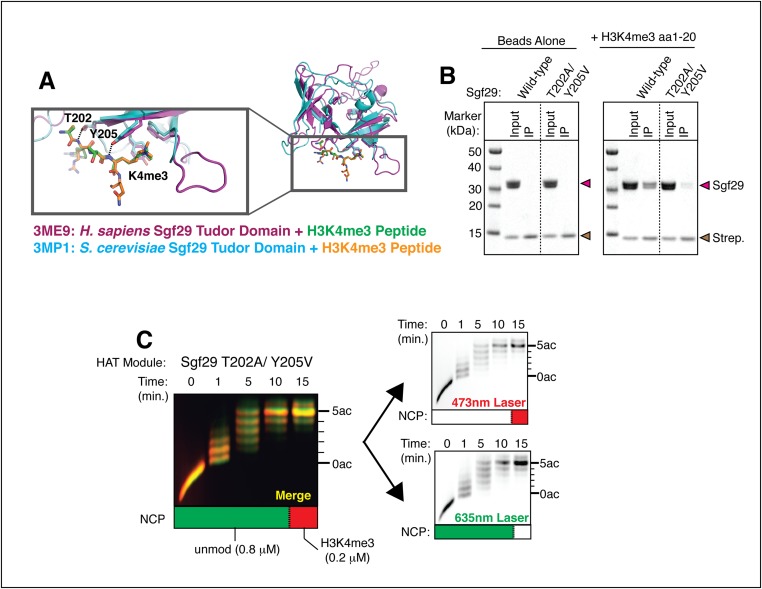
Fig. S5.
Mutations that disrupt binding to H3K4me3 by Sgf29 eliminate preferential acetylation of methylated nucleosomes. (A) Superimposed structures of the tandem Tudor domains from human (magenta) and yeast (blue) Sgf29 bound to H3K4me3 peptides (orange and green). T202A and Y205V mutations in Sgf29 eliminate hydrogen-bonding interactions with the histone H3 backbone. Mutating Y205 to valine also disrupts the aromatic cage within the methyl lysine-binding pocket while preserving local hydrophobicity. (B) H3K4me3 peptides pull down T202A/Y205V Sgf29 less efficiently than wild-type Sgf29. (C) Fluorescent images of a HAT reaction using the T202A/Y205V HAT module containing a 4:1 molar ratio of differentially labeled unmodified versus H3K4me3 nucleosomes resolved on an acid–urea gel. (Left) Merged image with unmodified nucleosomes colored green and H3K4me3 nucleosomes colored red. (Right) Individual images used to generate the merged image.