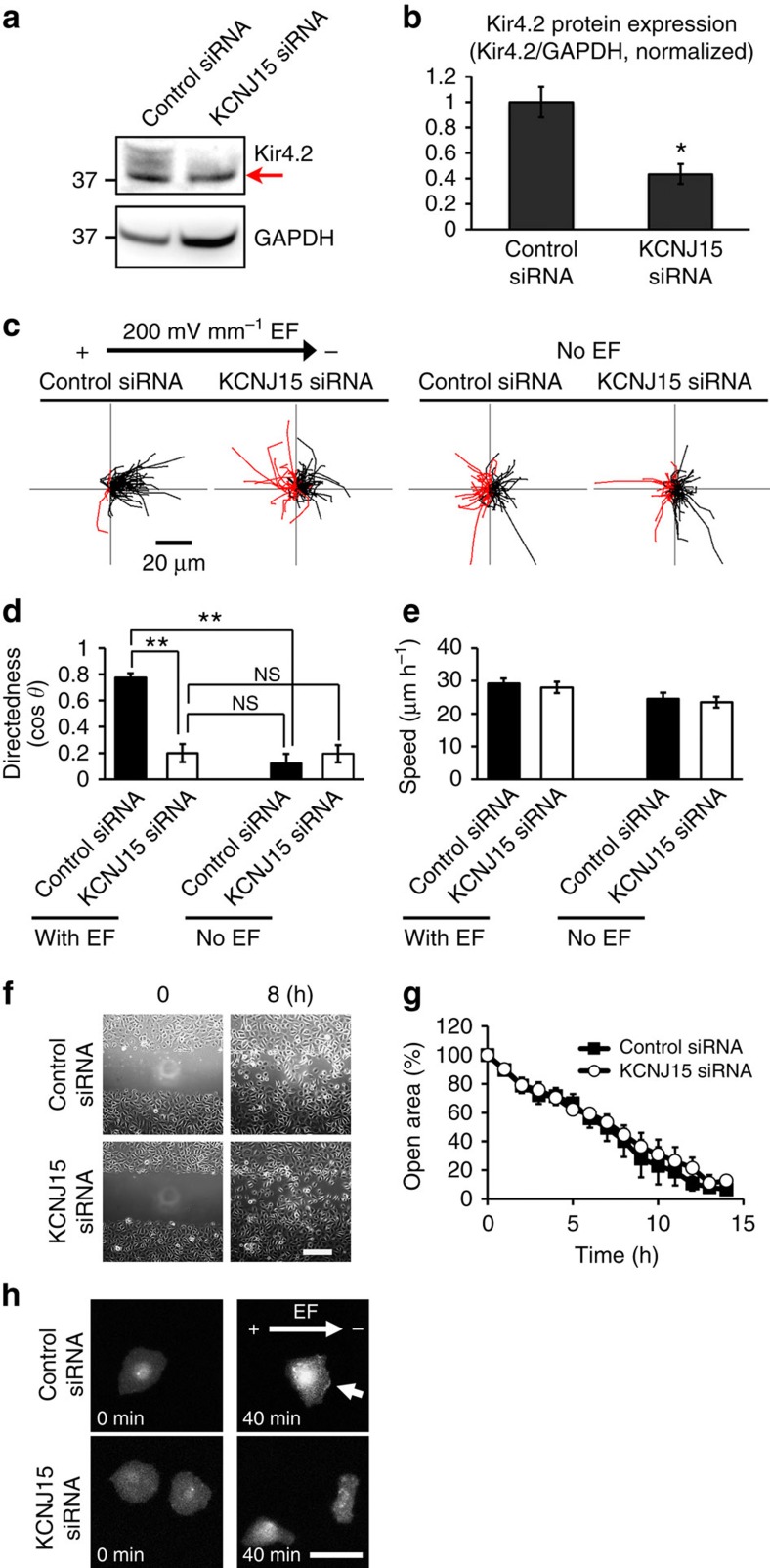
Figure 2. KCNJ15 knockdown specifically abolished galvanotaxis.
(a,b) Efficient knockdown of KCNJ15 shown with Western blotting, red arrow pointing to a non-specific band. Kir4.2/GAPDH ratio is used to quantify the protein level. n=3. (c,d) Migration trajectories and quantification of directional migration (directedness values (cos θ)) demonstrated that KCNJ15 knockdown abolished galvanotaxis and cells completely lost migration direction in an EF. Black and red lines indicate trajectories of cells migrated toward cathode and anode side, respectively. n=100 cells for each group, confirmed in two other replicates. (e) KCNJ15 knockdown did not affect cell migration speed whether in an EF or not (compare the trajectories in c). There are no statistically significance between each group. n=100 cells for each group, confirmed in two other replicates. (f,g) Directional cell migration in scratch assay were identical between KCNJ15 knockdown and scrambled RNAi treatment. Wound closure is represented as % of open area. When the error bars are not seen, the bars are smaller than the symbols. Wound was made using a pipette tip. There are no statistically significance between two groups. n=3. Scale bar in f, 200 μm. (h) KCNJ15 knockdown abolished cathodal distribution of Akt-PH-EGFP, a reporter for PIP3 localization. hTCEpi cells were transfected with siRNA and pcDNA3-Akt-PH-EGFP plasmid DNA. Fluorescence of Akt-PH-EGFP was recorded by fluorescence microscope. Arrow indicates PIP3 accumulation in cathode-facing side of control cells. Scale bar, 50 μm. Cells were transfected with siRNA against KCNJ15 or control oligo, and incubated for 48 h. EF=200 mV mm−1. Statistical analyses were performed by Student's t-test. Data represented as mean±s.e.m. *P<0.05; **P<0.01. NS, not significant.