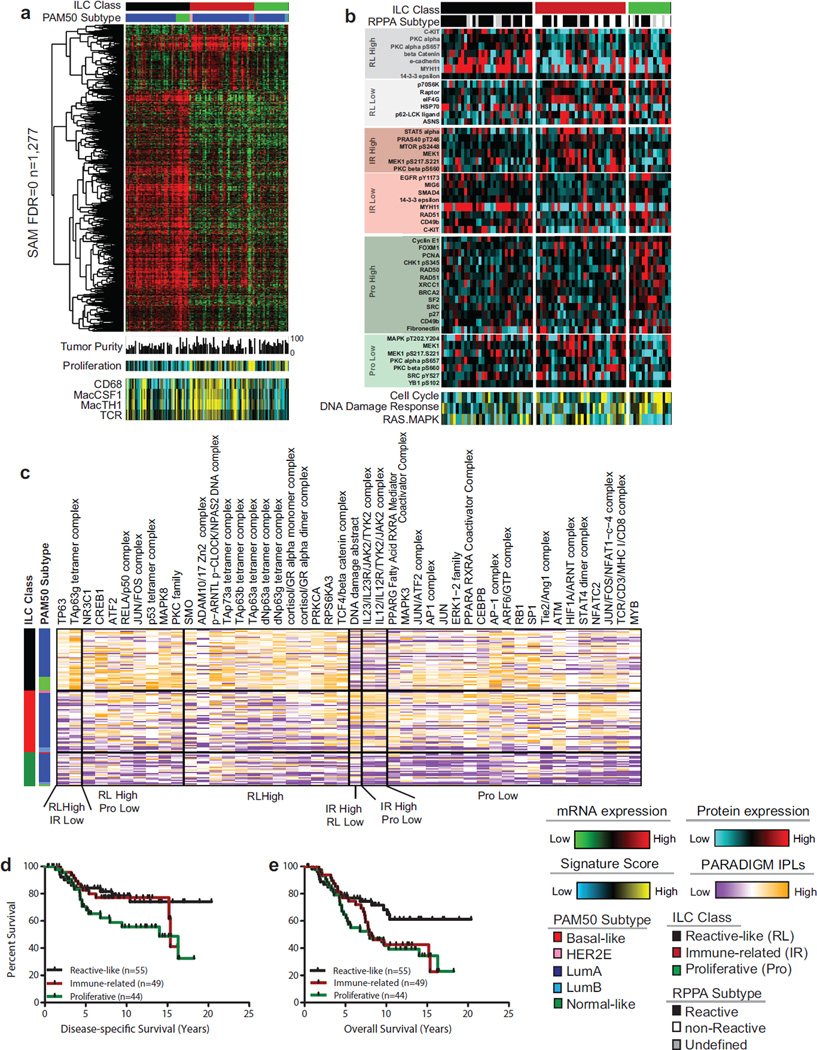
Figure 5. ILC molecular subtypes.
A) Three molecular subtype of lobular breast cancer were identified based on differential gene expression and show unique patterns highly expressed genes (n=1277, SAM FDR=0, upper panel), minor difference in tumor purity measured by ABSOLUTE, and differences in gene expression signatures measuring proliferation, CD68, Macrophage-associated CSF1, Macrophage–associated TH1, and T Cell Receptor Signaling (lower panel). Proliferation is highest in the proliferative (Pro) and immune-related (IR) subgroups; macrophage associated signaling is highest in immune-related tumors. B) Differences in protein expression profiles as determined by RPPA analysis. The reactive-like (RL) subgroup shows a significant association (p<1E-4, Fisher’s Exact test) with the RPPA-defined Reactive subgroup of breast cancer. Differences in subgroup-specific patterns of protein expression (p<0.05, t-test) for individual proteins (upper panel) as well as for protein expression signatures (lower panel) were identified. The proliferative subgroup shows higher expression of the cell cycle and DNA damage response pathways and lower levels of Ras-MAPK signaling (p<0.05). C) Subgroup-associated signaling features identified by PARADIGM. D) Reactive-like (n=55) tumors have significantly better disease specific (p=0.038, HR: 0.47, log-rank) and E) overall survival (p=0.022, HR=0.50) compared to proliferative (n=44) tumors in the METABRIC cohort. See also Figure S5.