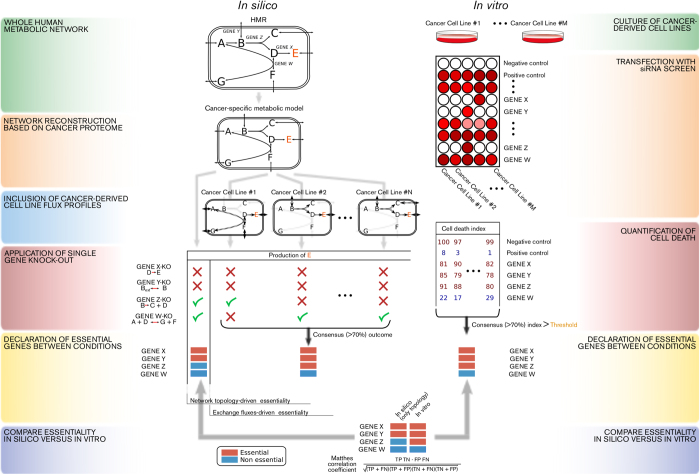
Figure 1. Strategy to measure the accuracy of flux balance analysis predictions of gene essentiality in cancer metabolism.
Left part: the Human Metabolic Reaction (HMR) database was used as a generic genome-scale metabolic network to reconstruct a cancer-type specific network based on proteome data obtained from cancer specimen (in the example, the reaction A fi B is absent in the cancer-specific model due to lack of the matched enzyme at the protein level for that cancer type). Successively, flux balance analysis is used to simulate whether a flux towards production of biomass (metabolite E) was feasible after every single gene-knockout, using as constraints either the topology of the cancer type-specific metabolic network or the measured fluxes for a number of exchange metabolites in different cancer type-derived cell lines. In the latter case a gene is deemed essential if it disables biomass production in ≥70% of the cell lines. Grey arrows indicate reactions not occurring in the network. Dashed arrows indicate measured fluxes in a cell line. Right part: cancer-derived cell lines were cultured and transfected with a library of siRNAs that target ~230 metabolic genes and cell number was determined after 4 days. If ≥70% of the cell lines passed a given threshold of cell death, the corresponding gene was deemed essential. Bottom: gene essentiality for the ~230 genes targeted by the siRNA library was compared in silico vs. in vitro and the accuracy of the predictions was calculated by several statistical measures (e.g. the Matthews Correlation Coefficients).