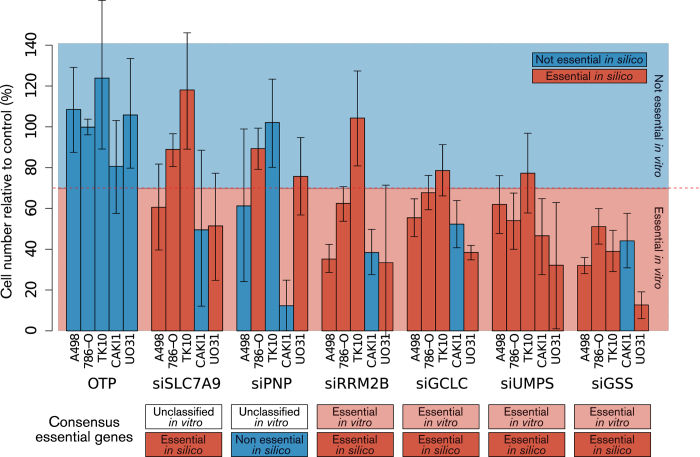
Figure 4. Validation of predicted gene essentiality in ccRCC.
Five ccRCC cell lines that match the flux profile constraints implemented to predict gene essentiality in ccRCC were transfected with siRNA targeting SLC7A9, PNP, RRM2B, GCLC, UMPS and GSS. A non-targeting oligonucleotide, OTP (scrambled siRNA), was used as negative control. . Each bar represents the mean cell number reduction relative to control together with the 95% highest density interval of two experiments performed in triplicate. The consensus outcome across cell lines in terms of gene essentiality is shown below each set of bars corresponding to a certain silenced gene. Genes that, if silenced, cause a ≥ 30% reduction in cell number relative to the non-targeting RISC-free siRNA in ≥ 70% of ccRCC cell lines are deemed essential in vitro. The consensus outcome for each silenced gene is compared to the prediction of essentiality in silico for the corresponding gene in ccRCC (compare with Fig. 3A).