Fig. 5.
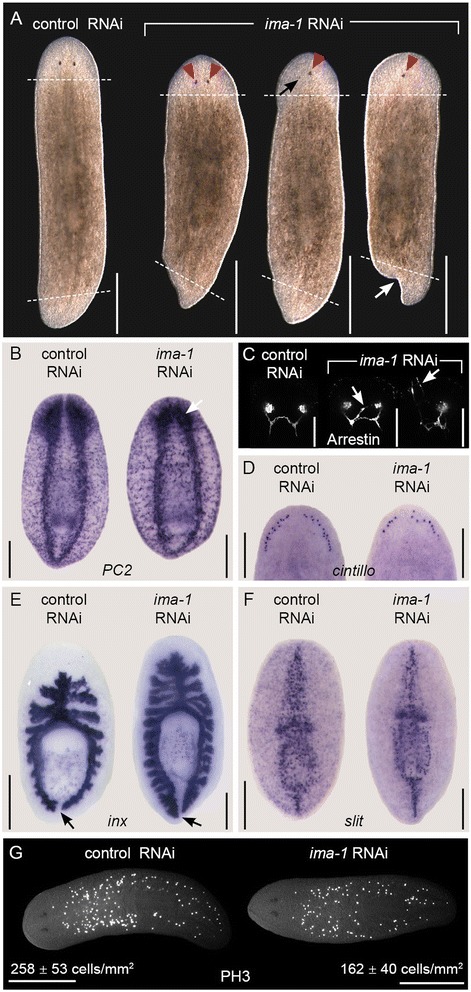
Smed-ima-1 is required for normal stem cell function and regeneration. a-f Images of control and Smed-ima-1(RNAi) animals fed bacterially expressed dsRNA targeting each gene four times over 2 weeks then amputated transversely both pre- and post-pharyngeally. Animals were imaged or fixed on day 10 of regeneration. Anterior is toward the top. Live animals were imaged from the dorsal side and all others were imaged ventrally. Scale bars = 0.5 mm for (a) and (g), 0.25 mm for (b) and (d-f), 0.1 mm for (c). a Live animals following RNAi treatment. Dashed lines indicate amputation sites. Red triangles indicate photoreceptors forming abnormally close to the midline, and the black arrow indicates a mis-positioned and underdeveloped photoreceptor. The white arrow indicates forking of the tail blastema. b In situ hybridization to the neuronal marker Smed-pc2. White arrow indicates small cephalic ganglia collapsed toward the midline. c Staining with anti-Arrestin antibody (Arrestin) to mark photoreceptor neurons. Arrows indicate aberrant neuronal projections. d In situ hybridization to Smed-cintillo, which labels sensory neurons. e In situ hybridization to Smed-inx to label the intestine. Arrows mark space between the two posterior intestinal branches. f In situ hybridization to midline marker Smed-slit. g Uninjured animals stained with anti-phospho-Histone H3 antibody (PH3) to mark mitotic cells following six dsRNA feedings. Anterior is to the left
