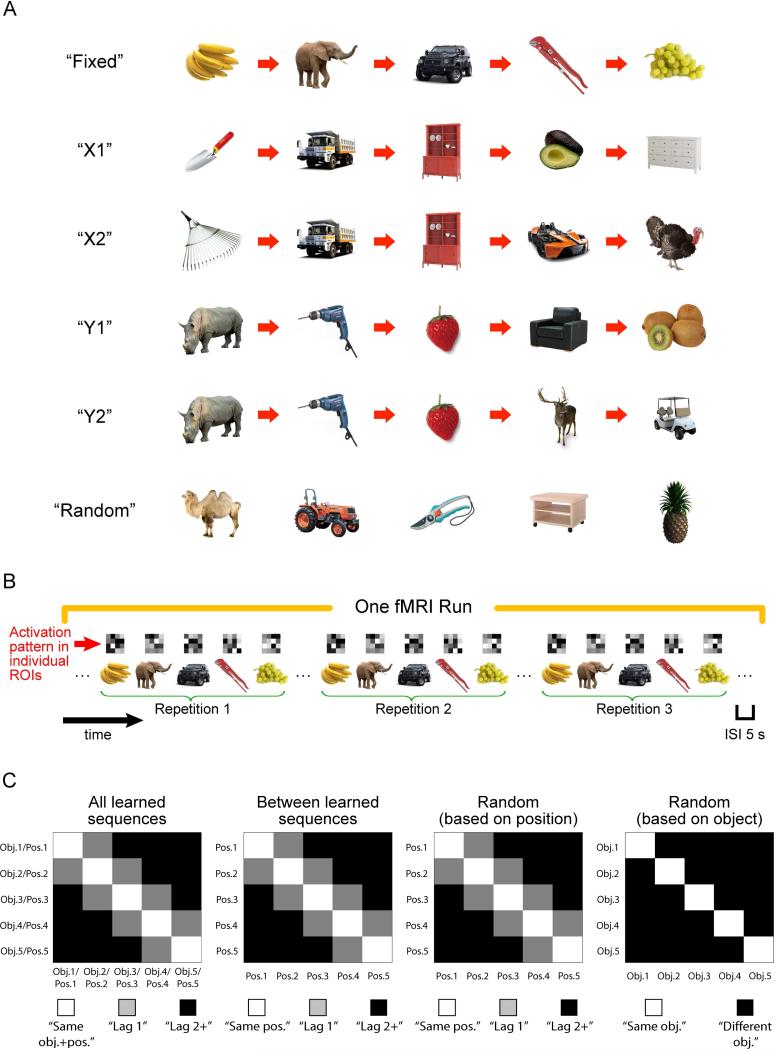
Figure 1.
Illustration of object sequences, sequence retrieval, and pattern analysis. A. The six types of objects sequences used in the current study. Each sequence consisted of five distinct objects organized in a specific order, except for the “Random” sequence in which the temporal order between the five distinct objects was randomly determined on each repetition. As a result, participants could not associate an object with a specific serial position in the “Random” sequence. B. Schematic of object sequence retrieval. Each object sequence was repeated three times in an fMRI run (five runs in total). The order in which object sequences appeared was pseudo-randomized such that there was no back-toback repetition of an object sequence and all six object sequences must have been presented before subsequent repetitions. The green brackets below the object sequence are to highlight repetitions of an object sequence in an fMRI run. In the real experiment, participants simply saw a continuous stream of objects without explicit cues to mark divisions between object sequences. Inter-stimulus-intervals (ISIs) were kept constant across trials (i.e., 5 s) throughout retrieval of object sequences. The matrix shown above each trial is to depict hypothetical multivoxel activation pattern associated with an object trial within an anatomically defined ROI. C. Pattern analyses associated with learned and “Random” sequences. Pattern similarity was computed by correlating multivoxel activity patterns between every possible pair of trials between repetitions of an object sequence (see also Methods for details), and could be summarized by a 5 by 5 similarity matrix. Similarity matrix for “All learned sequences” was computed by averaging together five 5 by 5 similarity matrices, each of which was associated with a learned, constant object sequence (i.e., “Fixed”, “X1”, “X2”, “Y1”, and “Y2”). Diagonal elements of the similarity matrix for “All learned sequences” reflected pattern similarity across repetitions of the same objects in the same serial position (i.e., “same obj.+pos.”). Offdiagonal elements reflected pattern similarity between trials that didn't share the same object information and were one position (i.e., “lag 1”) or two or more than two positions apart (i.e., “lag 2+”). Similar analysis procedures were applied to compute pattern similarity across repetitions of different learned sequences (excluding X1, X2 and Y1, Y2 similarity matrices). However, because each learned sequence consisted of a distinct set of objects, diagonal elements only reflected pattern similarity between trial pairs that shared the same position information (i.e., “same pos.”). Coding of position information could also be accessed by correlating between repetitions of the “Random” sequence (“Random” sequence (i.e., “Random (based on position)”). Because each serial position was not associated with a specific object in the “Random” sequence, similar to the analyses across learned sequences, diagonal elements only reflected pattern similarity between trial pairs that share the same position information (i.e., “same pos.”). Data associated with the “Random” sequence could also be rearranged, such that diagonal elements of the similarity matrix were computed from repetitions of the same objects but occupied different serial positions (i.e., “same obj.” in “Random (based on object)”).