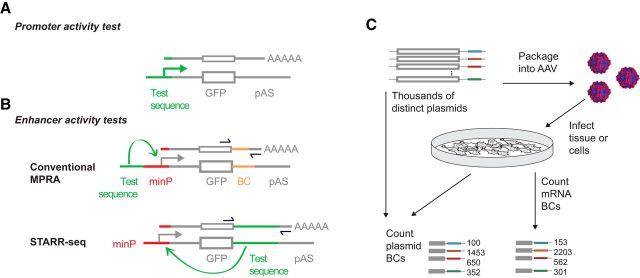
Figure 2.
Detecting enhancer activity using reporter assays and massively parallel reporter assays (MPRAs). A, Promoter activity is detected by fusing a test sequence upstream of a reporter sequence (i.e., GFP) and introducing it into cells or embryos. B, Enhancer activity is detected by fusing a test sequence near a minimal promoter and reporter. Alternatively, enhancer activity may be detected with the test sequence in the 3′ untranslated region (UTR) of a reporter gene (the STARR-seq configuration; Arnold et al., 2013; Cotney et al., 2015). For high-throughput analysis of enhancer activity, identifying sequences (such as barcodes, BCs) are placed in the 3′ UTR to identify individual enhancers so that they may be combined for “massively parallel” or multiplex quantification. C, MPRAs rely on libraries of plasmids in which the enhancer activity of each test sequence is quantified through sequencing of the BCs (or for STARR-seq, the enhancer itself). For MPRAs in primary neurons or in vivo, enhancer libraries can be packaged into AAV and transduced.