Figure 1. The process of ubiquitination consists of the sequential and cooperative actions of three classes of enzymes that catalyze the attachment of the protein Ubiquitin (Ub, green spheres) to substrate proteins.
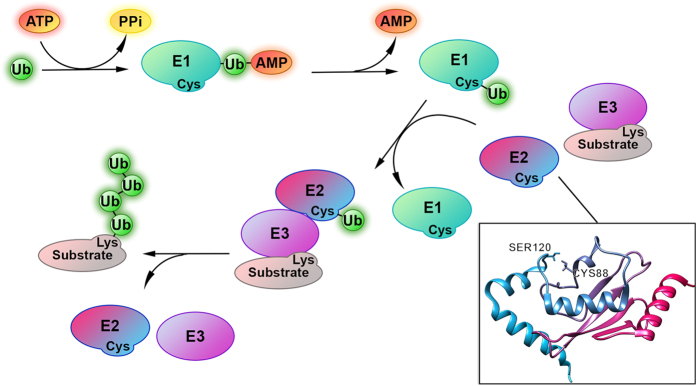
Ub is first activated by ubiquitin-activating enzyme (E1, green ovals) by an ATP-dependent reaction. The Ub-AMP product remains bound to the E1. Then a cysteine residue attacks the Ub, leading to an E1 ~ Ub thioester intermediate. Subsequently the Ub is transferred to a conserved cysteine in the active site of an ubiquitin-conjugating enzyme (E2, hot pink-azure ovals). The E2s catalyze the transfer of Ub to a lysine on the substrate (pink-grey ovals) to form an isopeptide bond, together with an ubiquitin ligase (E3, purple ovals). Alternatively, E2s can transfer Ub to a catalytic cysteine on HECT family E3s, which then transfer Ub onto substrate to form an isopeptide bond. E3s are important for recognition and selection of the substrate. Certain E2/E3 pairs catalyze several rounds of Ub attachment to generate substrates with poly-Ub chains. Substrate can be mono or poly-ubiquitinated and these modifications mark the substrate for different destiny and functions. In the bottom-right panel the structure of Human Ubiquitin-conjugating enzyme E2 B (PDB ID 1JAS, hHR6B) is showed as cartoon. The catalytic cysteine (Cys88) present in the Ubiquitin Conjugation (UBC) domain of E2, that forms thioester bond with Ub, and the serine (Ser120) in the conserved E2 Ser/Asp site (CES/D), whose phosphorylation enhances E2 catalytic activity, are highlighted as sticks.
