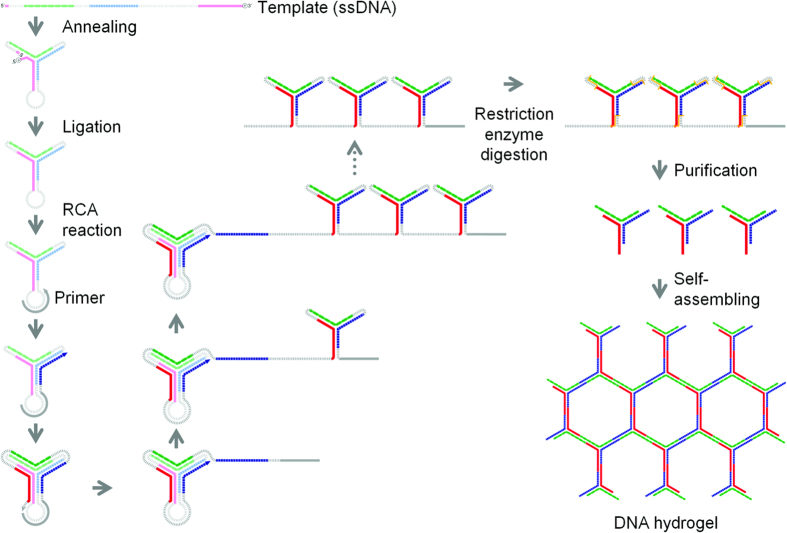
Figure 1. Schematic diagram of the mass amplification of tripodna with adhesive 5′-ends.
The template oligodeoxynucleotides (template 1) were designed to satisfy the following requirements: (a) the polypodna automatically forms by self-assembly; (b) each pod of the polypodna contains a 9 base long TspRI restriction digest site; (c) Each 5′-terminal end is phosphorylated in order to ligate with 3′-terminal within the polypodna body, and (d) connecting chain is added to the 3′-terminal of the polypodna to allow polypodna to be connected to one another. The designed templates were amplified via the following steps. (1) The template ssODNs were circularized using T4 DNA ligase. (2) After annealing the primer (primer 1), the DNA template was amplified through rolling circle amplification technique using Phi29 polymerase. (3) Before enzyme digestion, the RCA product was treated with EDTA and folded. (4) Long single-stranded DNAs were digested using restriction enzyme. (5) The target sequences were purified by size chromatography. (6) The resultant DNAs self-assembled after annealing, and they formed a hydrogel.