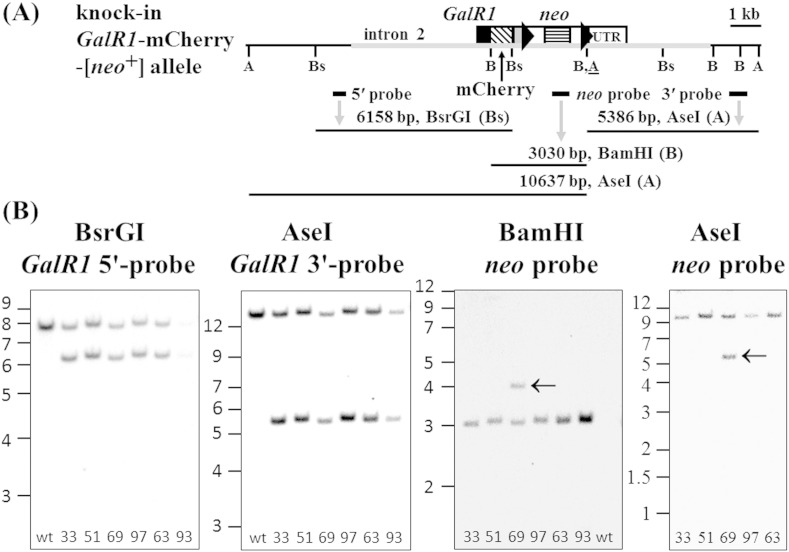
Supplementary Fig. 1.
Southern blot analysis of ES cell clones validates correct insertion of the GalR1-mCherry-[neo+] targeting construct. (A) Schematic diagram of the knock-in GalR1-mCherry-[neo+] allele showing the relative locations of: the targeting construct (grey horizontal thickened line); GalR1 exon 3 coding sequence (CDS; black filled box); mCherry CDS (diagonal banded box); the heterologous 3′-UTR (grey filled box); downstream FRT sites (right arrowheads) flanking an SV40-neo cassette selection marker; the endogenous 3′-UTR (UTR); restriction sites AseI (A), BsrGI (Bs), BamHI (B) and an introduced AseI restriction site immediately downstream of the 3′ FRT site (A); external 5′ and 3′ probes, neo probe, and hybridizing DNA fragments (see main article, Fig. 1A, for corresponding diagram of the endogenous allele). (B) DNA from six potential heterozygous GalR1-mCherry-[neo+] knock-in ES cell clones (33, 51, 63, 69, 93 and 97) and control wild-type mouse tail (wt) were digested and hybridized with: BsrGI and GalR1 5′ external probe (endogenous 7821 bp and knock-in 6158 bp; under-exposure for clone 93); AseI and GalR1 3′ external probe (endogenous 12,924 bp and knock-in 5386 bp); BamHI and neo probe (knock-in 3030 bp); or AseI and neo probe (knock-in 10,637 bp). The relative distance travelled by DNA ladder fragments (1 kb Plus, Life Technologies) are indicated in kb on the left of each panel. Neo was not detected in control wild-type mouse tail DNA, but an additional insertion of neo-hybridizing DNA was detected in ES cell clone 69 (arrowed) that was distinguishable from the correctly inserted knock-in fragment by digestion with BamHI or AseI. This additional neo insertion is due to only a fragment of introduced DNA (Fig. 1A, a grey horizontal thickened line), as the neo-hybridizing portion of AseI-digested DNA must be of at least 7366 bp which is larger than the additional hybridizing band of approximately 5500 bp, and quantitative genomic PCR detected only one copy of GalR1-mCherry.