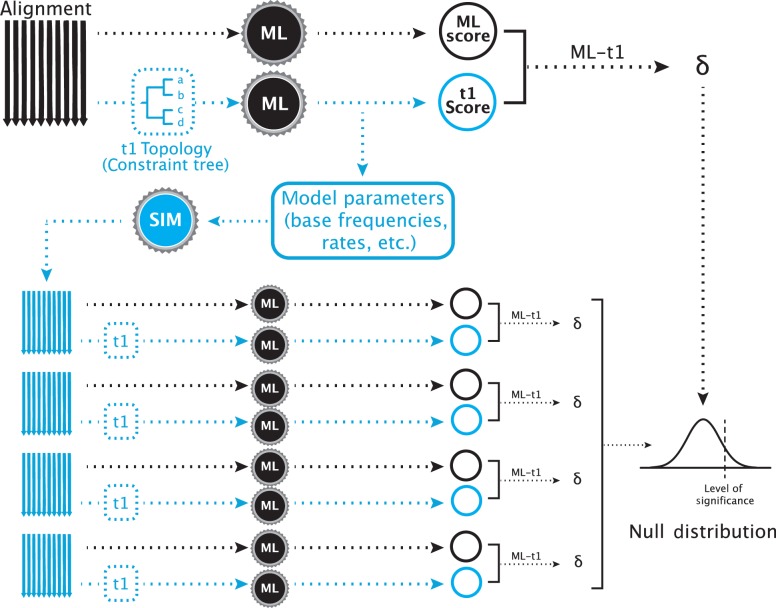
Figure 1.
A typical SOWH test. The test begins with two maximum likelihood searches on a single alignment. One search, represented by the black arrow, is performed with no constraining topology. Another test, represented by the shaded arrow, is constrained to follow an a priori topology that represents a phylogenetic hypothesis incongruent with the maximum likelihood topology. The black gears represent maximum likelihood software used to score the trees (i.e., GARLI, RAxML). These two searches result in two maximum likelihood scores, the difference () between which is the test statistic. From the constrained search, the optimized parameters and topology are retrieved and used to simulate new alignments with software (shaded gear) such as Seq-Gen. For each simulated alignment (shaded), two maximum likelihood searches are performed, one unconstrained (black arrow) and one constrained (shaded arrow), scores are obtained, and a value is calculated. The test statistic is compared to this distribution of values. A significantly large value is one which falls above some proportion of those generated by data simulation (i.e., 95%)