Figure 2.
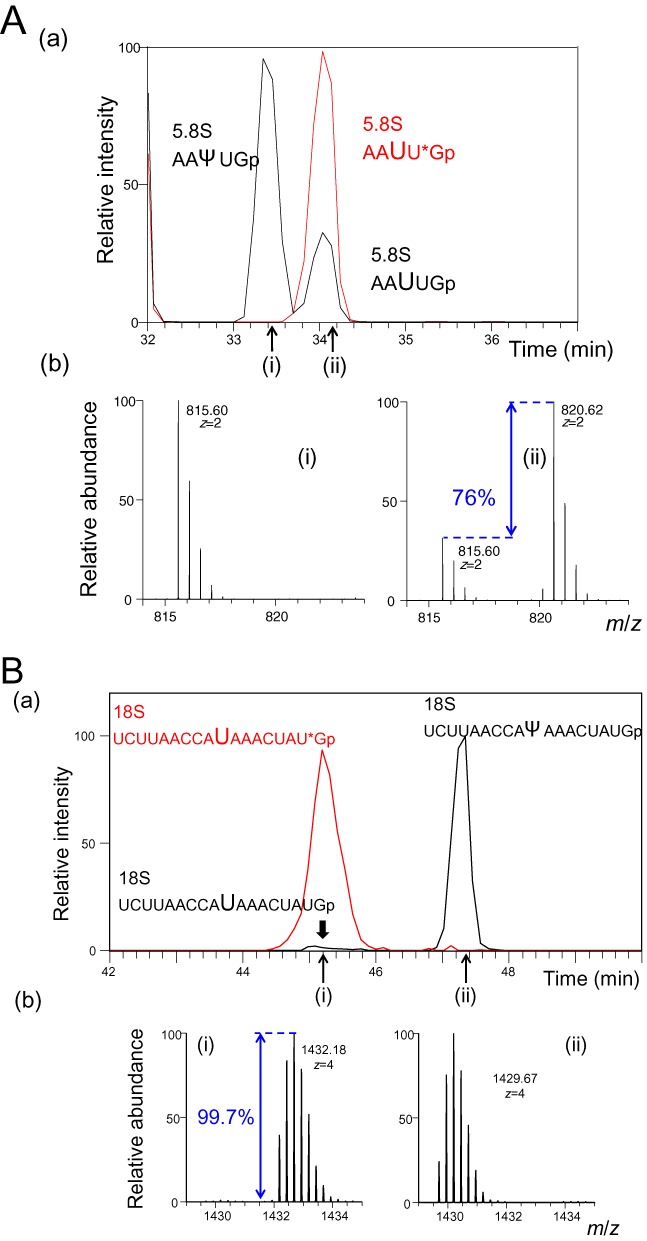
SILNAS-based PTM analysis. (A) Ψ76 in S. pombe 5.8S rRNA. (a) Extracted ion monitoring of RNase T1 fragments of 5.8S rRNA containing PTMs. Because PTM of U76 was incomplete (76% as estimated by SILNAS), RNase T1 produced two fragments, 74AAΨU78Gp and 74AAUU78Gp, from the sequence region 74–78 of 5.8S rRNA. In a subsequent LC analysis, the fragment 74AAΨU78Gp eluted earlier than the unmodified fragment 74AAUU78Gp and the corresponding heavy fragment 74AAUU78*Gp (*G, 13C10-guanosine) derived from the reference RNA. Oligonucleotide ion masses: [AAUUGp]2− and [AAΨUGp]2− (black line), m/z 815.60; [AAUU*Gp]2− (red line), m/z 820.62. (b) MS spectrum for each RNA peak, (i) and (ii). From the signal heights of MS spectra of the light and heavy AAUUGp ion, the extent of PTM was estimated as 76%. (B) Ψ1039 in S. pombe 18S rRNA. (a) Extracted ion monitoring of RNase T1 fragments of 18S rRNA containing PTMs. Although PTM of U1039 was almost complete (99.7% as estimated by SILNAS), a trace amount of unmodified RNase T1 fragment, 1030UCUUAACCAUAAACUAU1047Gp, was detected with the modified fragment 1030UCUUAACCAΨAAACUAU1047Gp from the sequence region 1030–1047 of 18S rRNA. In a mass chromatogram, the modified fragment 1030UCUUAACCAΨAAACUAU1047Gp eluted later than the unmodified fragment 1030UCUUAACCAUAAACUAU1047Gp and the corresponding heavy fragment 1030UCUUAACCAUAAACUAU1047*Gp (*G, 13C10-guanosine) derived from the reference RNA. Oligonucleotide ion masses: [UCUUAACCAUAAACUAUGp]4− and [UCUUAACCAΨAAACUAUGp]4− (black line), m/z 1429.67; [UCUUAACCAUAAACUAU*Gp]4− (red line), m/z 1432.18. The mass windows used for extraction were 15 ppm. (b) MS spectrum for each RNA peak, (i) and (ii). From the signal heights of MS spectra of the light and heavy UCUUAACCAUAAACUAUGp ion, the extent of PTM was estimated as 99.7%.
