Figure 6.
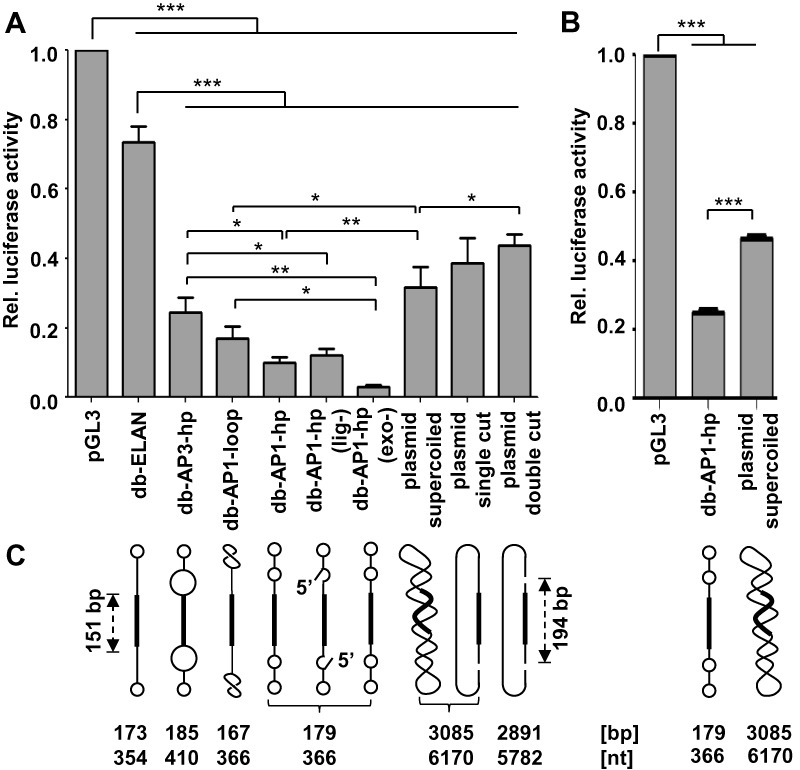
Luciferase target gene knockdown in HEK293T cells triggered by shRNA expressing dumbbells and plasmids. (A) Cells were co-transfected with 90 ng luciferase reporter vector pGL3 and 90 ng (equimass amounts) dumbbell or plasmid DNA. Firefly luciferase expression levels relative to the uninhibited negative control. Values are mean values ± SEM of two (db-ELAN) or three (rest) independent experiments. (B) Cells were co-transfected with 90 ng luciferase reporter vector pGL3 and 0.5 pmol (equimolar amounts) dumbbell or plasmid DNA. Firefly luciferase expression levels relative to the uninhibited negative control. Values are mean values ± SEM of three independent experiments. A and B, the statistical analysis was performed using repeated one-way ANOVA plus a post-hoc Newman–Keuls test. The significance was denoted as ***p < 0.001; **p < 0.01; *p < 0.05. (C) Structures/topology of the DNA vectors tested in A and B. All vectors harbor a 151 bp shRNA expression cassette. Hairpin loops and internal loops within the dumbbell (db) vectors are indicated as circles, i.e. small/large circles indicate small/large loops. All dumbbell vectors were tested after ligation and exonuclease treatment. Vector db-AP1-hp was additionally tested after skipping either the exonuclease treatment (exo-) or both ligation and exonuclease treatment (lig-). The pSuper plasmid vector was tested as supercoiled DNA, after linearization with KpnI (single cut), or after KpnI/BamHI digestion (double cut). The shRNA expression cassette was contained in the smaller 194 bp KpnI/BamHI fragment. AP1 or AP3 abasic site mimics were counted as 1 or 3 nt, respectively.
