Abstract
Smad proteins are important intracellular mediators of TGF-β signalling, which transmit signals directly from cell surface receptors to the nucleus. The MH1 domain of Smad plays a key role in DNA recognition. Two types of DNA sequence were identified as Smad binding motifs: the Smad binding element (SBE) and the GC-rich sequence. Here we report the first crystal structure of the Smad5 MH1 domain in complex with the GC-rich sequence. Compared with the Smad5-MH1/SBE complex structure, the Smad5 MH1 domain contacts the GC-rich site with the same β-hairpin, but the detailed interaction modes are different. Conserved β-hairpin residues make base specific contacts with the minimal GC-rich site, 5′-GGC-3′. The assembly of Smad5-MH1 on the GC-rich DNA also results in distinct DNA conformational changes. Moreover, the crystal structure of Smad5-MH1 in complex with a composite DNA sequence demonstrates that the MH1 domain is targeted to each binding site (GC-rich or SBE) with modular binding modes, and the length of the DNA spacer affects the MH1 assembly. In conclusion, our work provides the structural basis for the recognition and binding specificity of the Smad MH1 domain with the DNA targets.
INTRODUCTION
The transforming growth factor-β (TGF-β) signaling pathway regulates diverse biological processes including embryonic development and tissue homeostasis (1,2). Ligands of the TGF-β superfamily can bring together two different types of receptor Ser/Thr kinases (type I and type II receptors) on the cell-surface, resulting in their oligomerization and activation (3,4). Type I receptor kinases then activate their downstream intracellular transcription factor Smad proteins, resulting in ligand-induced transcription (5). As the central player of the canonical TGF-β signaling pathway, Smad proteins can be classified into three subclasses in vertebrates: the receptor-regulated Smads (R-Smad, including Smad1, 2, 3, 5 and 8), the common-mediator Smad (Co-Smad, Smad4) and the inhibitory Smads (I-Smad, Smad6 and 7) (6). R-Smads and the Co-Smad share two conserved domains connected by a proline-rich linker: the Mad homolog domain 1 (MH1), which is essential for specific DNA binding; and the Mad homolog domain 2 (MH2), which is often responsible for protein-protein interactions (7,8).
The ligands of the TGF-β superfamily can be divided into two major groups, TGF-βs and BMPs (bone morphogenetic proteins) (9). TGF-βs often induce the activation of Smad2 and Smad3, whereas Smad1, 5 and 8 predominantly respond to BMPs. The activation of R-Smad proteins requires the specific phosphorylation of the ‘SXS’ motif at the C terminus of the MH2 domain by type I receptor kinases, which is necessary for the formation of heterotrimers containing two phosphorylated R-Smads and one Smad4 molecule (10,11). Phosphorylated Smad complexes are then translocated into the nucleus, and disparate Smad complexes are selectively recruited to target genes (Supplementary Table S1). The Smad complex formed by activated Smad2/3 and Smad4 proteins mostly binds to the 5′-GTCT-3′ motif (the Smad binding element, SBE) (12–14). Many TGF-β-responsive promoter regions contain one or multiple copies of SBEs and tandem repeats of SBE can confer TGF-β inducibility (13–16). The presence of multiple SBEs might increase the binding affinity through the cooperative interactions of the MH1 domains in the Smad complex. In contrast, the Smad1/5/8-Smad4 complex prefers GC-rich sites flanked by SBE motifs in vivo (17–21). The first GC-rich sequence, GCCGnCGC, was originally identified in Drosophila (22). In mammals, similar GC-rich sequences, such as GCCG and GGCGCC, have been evaluated in the promoter regions of several BMP target genes and further studies have identified a Smad1/5 binding motif, the GC-rich Smad binding element (GC-SBE) (19–21,23). The difference in the DNA binding specificities of the TGF-β and BMP Smad complexes contributes to different ligand-induced gene expressions, such as antagonism between the two types of ligands in some biological processes (24–26).
As the MH1 domain of Smad is responsible for direct DNA contact, understanding how the MH1 domain recognizes DNA and how R-Smads–Smad4 complexes bind to different DNA sequences is essential for delineating Smad transcriptional programs. The first Smad-MH1/DNA complex structure was that of the TGF-β-regulated Smad3 MH1 domain and a DNA sequence containing palindromic SBE sites (27). A β-hairpin formed by β2 and β3 strands of the MH1 domain is inserted into the major groove of the DNA and establishes hydrogen bonds with nucleotides in the SBE site. This mode of interaction is also conserved in recognition of SBE DNA by the Smad1 (BMP Smad) and Smad4 (Co-Smad) MH1 domain, validated by crystal structures (28,29). However, in vitro DNA binding assays of Smad1/3/4 demonstrated that these three Smad MH1 proteins possess different affinity and cooperativity for binding to the palindromic SBE DNA (28,29). Similarly, in the case of GC-rich DNA binding, EMSA assays were performed using the Smad1/3/4 MH1 domains in complex with a ‘GGCGCC’-containing DNA (28,29). The Smad4 MH1 domain formed rather unstable monomeric complexes, while both the Smad1- and Smad3-MH1 domains were able to bind the DNA with a 2:1 ratio but with different cooperative binding patterns. However, due to the lack of direct structural evidences, the interaction mode of the MH1 domain when complexed with the GC-rich DNA, and the means by which Smad proteins discriminate between SBE and GC-rich DNA sequences remain elusive. Homology modeling of the BMP-responsive Smad proteins using the Smad/SBE structure as a template also failed to provide a solution to this problem (30). Further to this, although biochemical assays have also been performed in order to resolve the DNA recognition of Smad (17), the minimal binding module recognized by Smad-MH1 on the GC-rich sequence is still unclear and the exact pairings of amino acids and bases in base-specific DNA binding also need to be determined.
In order to understand in more depth the mechanism of DNA binding specificity of the Smad MH1 domain, we conducted biochemical and structural analyses on mouse Smad5-MH1. The electrophoretic mobility shift assays (EMSA) demonstrated that the Smad5 MH1 protein could recognize both SBE and GC-rich sites, and that it bound to the GC-rich sequences in a highly cooperative manner. In addition, we solved three structures of Smad5-MH1 in complex with different types of DNA elements: a palindromic SBE sequence, a palindromic GC-rich sequence (GC-BRE), and a composite DNA sequence containing both SBE and GC-rich sites (GCRj2). Among these structures, the Smad5 MH1 molecules recognize the two types of binding sites (SBE and GC-rich) using the same DNA recognition β-hairpin. However, the base-specific DNA bindings are provided by direct hydrogen bond interactions involving different pairings of amino acids and bases. The 3-bp 5′-GGC-3′ sequence represents the minimal GC-rich Smad binding site. Compared with the 4-bp SBE sequence, the compressed GC binding site would facilitate direct interactions between the adjacent bound MH1 molecules on repeated GC-rich sites, and would also induce different changes in the global and local shape of the DNA duplexes. Our structures unravel the mechanism of Smad-MH1 binding to the GC-rich site and the modular binding mode of the MH1 domains to multiple SBE or GC-rich sites. Therefore, this work provide a more detailed illustration of the DNA binding mechanism of Smad proteins, and the structural basis for an in-depth understanding of BMP and TGF-β signaling.
MATERIALS AND METHODS
Protein and DNA preparation
Plasmids of the full-length mouse Smad5 and the full-length human Smad3 were kindly provided by Dr. S.C. Lin (Xiamen University). The fragments of Smad5 MH1 (1–143) and Smad3 MH1 (1–145) were generated by standard PCR procedure and inserted into the pET21b and pGEX4T-1 vectors, respectively. Plasmids were transformed into the E. coli BL21(DE3) strain. The cells were grown at 37°C in Luria-Bertani (LB) medium to an OD600 of approximately 0.6. Protein expressions were then induced with 0.2 mM isopropyl-β-d-1 thiogalactopyranoside (IPTG) at 16°C for 14 h. Cells were harvested by centrifugation and lysed by sonication in buffer containing 50 mM Tris-HCl, pH 8.0 and 200 mM NaCl. The His6-tag fused Smad5 MH1 domain proteins were first isolated over Ni-NTA columns (Qiagen), while the GST-tagged Smad3 MH1 domain proteins were first purified over Glutathione-Sepharose 4B columns (GE Healthcare). The GST tag was removed by on-column cleavage using thrombin. The eluted proteins from the affinity columns were then subjected to cation exchange chromatography (SOURCE 15S, GE Healthcare) and eluted with linear NaCl gradient. Fraction containing the Smad MH1 proteins were concentrated by ultracentrifuge and then subjected to size-exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare) in a buffer containing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl and 5 mM dithiothreitol (DTT). All proteins were stored at -80°C and the protein purity was ≥95% as determined by SDS–PAGE. DNA oligonucleotides used for EMSA and crystallization were synthesized by Invitrogen. Single-stranded DNA was mixed with an equimolar amount of a complementary strand and annealed at 85°C by slow cooling to room temperature over a period of 5 hr. Double-stranded DNAs were stored at -20°C.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (28), with a few modifications. Briefly, freshly thawed Smad MH1 proteins were serially diluted, mixed with 1 μM dsDNAs and incubated at 4°C for 1 h. Then, 8 μl of the reaction mixture was loaded onto 10% native PAGE gels, electrophoresed using Tris-Glycine buffer (25 mM Tris, pH 8.3, 192 mM Glycine) at 180 V for 30 min at 4°C and the gel was stained with ethidium bromide.
Crystallography and structure determination
Protein-DNA complexes were prepared by mixing dsDNA with equimolar amounts of the Smad5 MH1 domain. All crystals were grown by the hanging-drop vapor diffusion method at 21°C. Crystals of Smad5-MH1/SBE complex were grown by mixing the protein-DNA complex with an equal volume of reservoir solution containing 15% PEG3350 and 0.2 M magnesium formate. Crystals of Smad5-MH1/GC-BRE complex were grown in a reservoir buffer containing 0.1 M Tris, pH 7.0, 12% PEG3350 and 0.2 M KNO3. Crystals of Smad5-MH1/GCRj2 complex were obtained by mixing the protein-DNA complex with an equal volume of reservoir solution containing 0.1 M Bis-tris, pH 6.5, 0.2 M LiNO3, 8% PEG3350 and 5% glycerol. All Crystals were equilibrated in a cryoprotectant buffer containing reservoir solution supplemented with 30% glycerol and then flash frozen in liquid nitrogen. All diffraction data sets were collected at beamline 17U at the Shanghai Synchrotron Radiation Facility and processed using the HKL2000 (31). The structures were solved by molecular replacement using Phaser with the Smad1 MH1 molecule in the Smad1/SBE complex (PDB id: 3KMP, chain A) as the search model (28,32). The ideal dsDNA was manually fitted to the strong electron density indicative of a DNA duplex in Coot (33). Further refinement was performed with Coot and PHENIX (34). All residues were in the allowed regions of the Ramachandran plot, as defined by MolProbity (35). The data processing and refinement statistics are summarized in Table 1. All structural representations in this paper were prepared with PyMOL (http://www.pymol.org). Topological DNA parameters were analyzed by program Curves+ (36).
Table 1. Crystallographic data collection and refinement statistics.
| Smad5-MH1/SBE | Smad5-MH1/GC-BRE | Smad5-MH1/GCRj2 | |
|---|---|---|---|
| PDB ID code | 4ZKG | 4ZL2 | 4ZL3 |
| Data Collectiona | |||
| Space group | P 21 21 21 | P 31 | P 31 |
| Cell dimensions | |||
| a, b, c (Å) | 71.54, 74.50, 83.74 | 92.73, 92.73, 83.61 | 119.46,119.46, 93.07 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 50.00–3.05 | 50.00–3.05 | 40.00–3.20 |
| (3.10–3.05)b | (3.14–3.05) | (3.31–3.20) | |
| Rmerge (%) | 7.7 (44.4) | 7.8 (59.6) | 9.5 (55.6) |
| I/σ (I) | 16.1 (4.7) | 13.8 (2.6) | 22.1 (4.5) |
| Completeness (%) | 89.9 (100.0) | 97.4 (100.0) | 98.8 (100.0) |
| Redundancy | 6.5 (7.2) | 4.6 (4.8) | 5.5 (5.7) |
| Refinement | |||
| Resolution (Å) | 34.04–3.05 | 46.37–3.06 | 29.94–3.20 |
| No. of reflections | 7863 | 14738 | 24192 |
| Rwork/Rfreec (%) | 23.0/26.2 | 25.0/27.4 | 24.5/28.9 |
| No. Atoms | |||
| Protein/DNA | 2624 | 4971 | 5744 |
| Zinc | 2 | 4 | 4 |
| Average B-factors | 85.7 | 120.9 | 123.9 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.005 |
| Bond Angles (°) | 0.932 | 1.053 | 0.913 |
| Ramachandran analysis (%) | |||
| Favored | 95.9 | 91.4 | 94.5 |
| Additionally allowed | 4.1 | 8.6 | 5.5 |
| Disallowed | 0 | 0 | 0 |
aAll data sets were collected from a single crystal.
bValues in parentheses are for the highest resolution shell.
cRfree was calculated on a random 5.0% reflections of the data.
RESULTS
Cooperative binding of Smad5-MH1 to specific DNA elements
It was previously reported that the BMP-regulated Smad1-MH1 domain exhibited a cooperative binding to the DNA, while the TGF-β-regulated Smad3 bound in an additive manner (28). To find out whether this mode of cooperative DNA binding mode is conserved in BMP-responsive Smad proteins, we performed gel mobility shift assays of the Smad5- and Smad3-MH1 proteins binding to either a palindromic SBE DNA or a palindromic GC-rich sequence (GC-rich BMP response elements, GC-BRE). In accordance with previous results, Smad3 bound additively to both the SBE and GC-BRE sequences. There were two clearly observed shifted bands in the gel, with the lower band representing one MH1 molecule bound to one DNA duplex and the higher band representing two MH1 domains bound to one DNA. However, Smad5-MH1 preferred to dimerize cooperatively on the palindromic DNA sequences since the 1:1 protein-DNA complexes only exist at a low level of protein concentration (Figure 1A and B). Therefore, both Smad5 and Smad1, which are BMP-responsive Smads, exhibit a different mode of DNA binding when compared with the TGF-β-responsive Smad3. Additionally, the Smad5 MH1 domain displayed a much stronger cooperative binding to the GC-BRE sequence than to the palindromic SBE sequence, in which the protein almost formed a constitutive dimer on the GC-BRE sequence (Figure 1B). This phenomenon indicated that there was an unknown mechanism by which two Smad5 MH1 molecules act cooperatively to assemble on the adjacent GC-rich sites. Sequence analyses of the BMP-responsive regions have also revealed that the GC-rich binding sites are often flanked with SBE elements (18–21,23,37–39). We therefore performed electrophoretic mobility shift assays to examine the binding pattern of Smad5 to the cis-regulatory sequences in BMP-responsive Id2 and Bambi, in which both GC-rich and SBE sites are identified and separated by variable length of spacer. Complexes of the Smad3 MH1 domain with DNA containing three repeats of the SBE motif were used as a molecular marker, as the additively binding pattern is adopted by Smad3 (Figure 1C and Supplementary Figure S1). Similar to the binding results for the GC-BRE sequence, Smad5-MH1 showed a highly cooperative assembly on the sequence of Id2 and Bambi, in which the band representing the MH1 monomer-DNA complex is barely visible in the gel. These data suggested that the binding mechanism of the Smad5 MH1 domain to diverse DNA sites might be different from that of the structural characterized Smad proteins. Although the amino acid sequence is highly conserved across the R-Smads (Figure 1D), the cooperative DNA binding mode is favored by Smad5.
Figure 1.
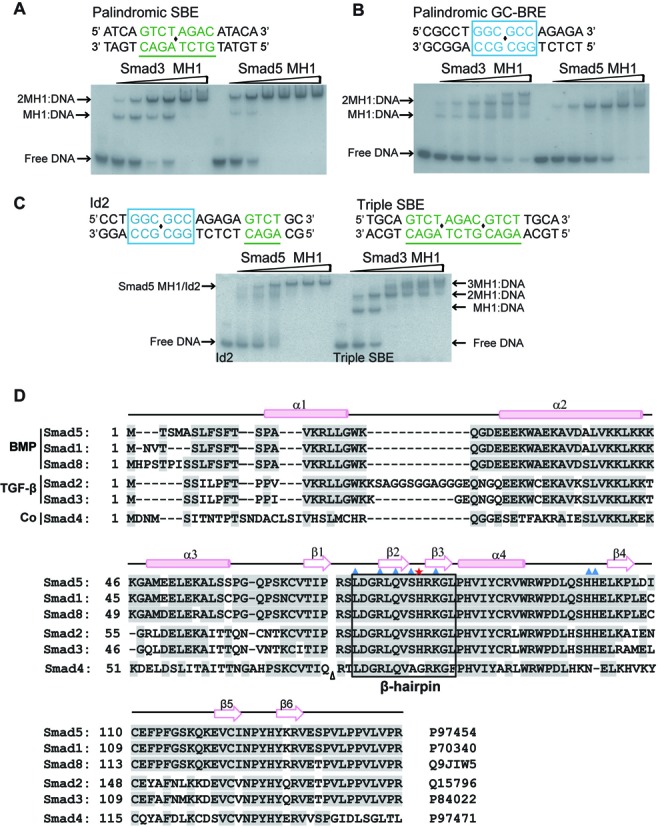
Cooperative binding of Smad5-MH1 to the DNA sequence. (A and B.) Binding of the Smad3 MH1 domain (left panel of the gel) and the Smad5 MH1 domain (right panel of the gel) to (A) palindromic SBE DNA; and (B) GC-BRE DNA. The DNA sequence in each assay is shown above the gel, in which the SBE site are colored green and underlined by green line, while the GC-rich sites are colored in blue and highlighted in a blue box. The centre of the palindromic sequence is marked with a black diamond. Protein concentrations used in EMSA assays were 0, 0.5, 1, 2, 3, 4, 5 μM (from left to right in each panel of the gel) and the concentration of DNA is 1 μM. (C) Smad5-MH1 binding to the Id2 promoter element. The complexes of Smad3-MH1 binding to the Triple SBE DNA sequence (right panel of the gel) were used as molecular weight markers. The EMSA assays were performed as that in Figure 1A and B. Sequence derived from Id2 promoter (-2781/-2762) and the triple SBE DNA sequence are shown above the gel in which the SBE and GC-rich sites are displayed as that in Figure 1A and B. (D) Multiple sequence alignment of the Smad MH1 domains. Secondary structure elements as seen in the Smad5 structures are placed on the top of the alignment. The code following each sequence is the corresponding UniProt ID. Amino acids that are involved in the direct interactions with dsDNA are marked with a blue triangle, except His80 (Smad5 numbered) labeled with a red asterisk. The DNA recognition β-hairpin is boxed. For clarity, a 30 amino acid insertion of Smad2 was omitted, indicated by a triangle.
Classic interaction mode in the complex of Smad5-MH1 with palindromic SBE DNA
In order to understand the mode of cooperatively binding of Smad5-MH1 on the target DNA sequences, we attempted to determine the crystal structures of Smad5 in complex with various responsive DNA elements. Firstly, by using a DNA element derived from the previously reported Smad1-MH1-DNA complex (28), we solved the crystal structure of Smad5-MH1 in complex with a palindromic SBE DNA sequence. This structure was refined to a final Rfree value of 27.6% at 3.05 Å (Table 1). The final model contains two monomers of Smad5-MH1 bound to the palindromic 16-bp SBE DNA duplex in an asymmetric unit (Figure 2A). Two MH1 monomers are arranged around a non-crystallographic 2-fold axis and contact the same 4-bp Smad binding element on opposite faces of the DNA, similar to that observed in all reported Smad-MH1-SBE complex structures (27–29,40). Each Smad5 MH1 domain adopts a globular fold consisting of four α-helices (α1-α4) and six short β-strands (β1-β6) connected by loops (Figures 1D and 2A). A zinc ion was modeled into the core of the MH1 structure, coordinated by the conserved cysteine (Cys65, Cys110 and Cys122) and histidine (His127) residues. Compared with previous Smad-SBE complexes, Smad5-MH1 exhibit an ‘open’ structure with the helix α1 protruding into the body of a symmetry-related molecule, similar to Smad1 (Supplementary Figure S2A and S2B). Superimposing of the Smad5-MH1-SBE complex and the Smad1-MH1-SBE complex results in a Cα RMSD of 0.45 Å for 210 aligning residues.
Figure 2.
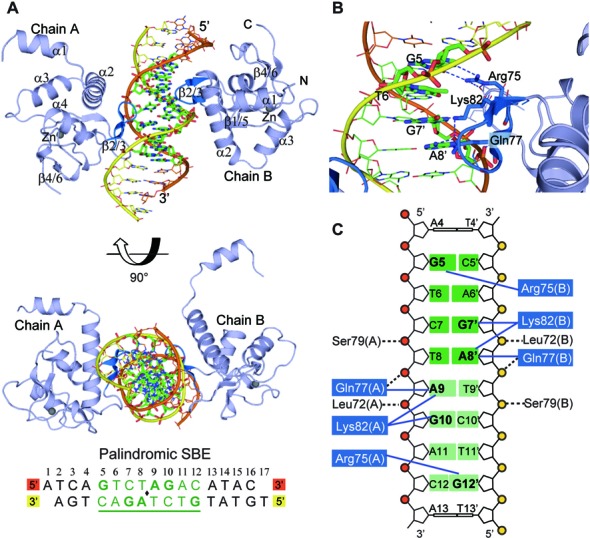
Crystal structure of Smad5-MH1 in complex with the palindromic SBE sequence. (A) Overall structure of the Smad5-MH1/palindromic SBE complex. The Smad5 MH1 domains are colored in light blue, with the β-hairpin highlighted in marine. The zinc atoms are shown as spheres. The template chain and the complementary chain of the dsDNA are shown in orange and yellow, respectively. The DNA bases of the central SBE site are shown as sticks and colored in green. The DNA sequence used for crystallization is shown below the structure, with the central 8-bp palindromic SBE site underlined and highlighted in green. (B) Base-specific interactions by the β-hairpin. Residues and DNA bases involved in specific binding are colored as marine and green sticks, respectively. Hydrogen bonds are represented by blue dashed lines. (C) Diagram summarizing Smad5-MH1/SBE interactions. Non-interacting nucleotides are omitted. Each copy of the SBE motif is shaded in dark green and light green, respectively. The conserved β-hairpin residues, Arg75, Lys82 and Gln77, are shaded in blue. Interactions between the amino acids and DNA bases are shown as blue solid lines, while the contacts with the DNA phosphates are shown as black dashed lines.
In the Smad5-MH1-SBE complex, each of the two MH1 molecules binds identically and independently to the major groove of a 4-bp Smad box, 5′-GTCT-3′ or 5′ -AGAC-3′. Compared with the structures of the Smad1- and Smad3-MH1 domains, the base-specific DNA contacts in Smad5 are conserved and virtually identical. The β-hairpin motif formed by β2 and β3 recognizes the SBE site, in which Arg75, Gln77 and Lys82 have contact with G5, G7’ and A8’, respectively (Figure 2B and C). Similar to Smad1, the ‘open’ N-terminus of Smad5 leads to the rearrangements at helix α2 and the α1/α2 hinge, which results in the impeding of several DNA contacts involving the helix α2 seen in Smad3 (Supplementary Figure S2B) (27,40). For instance, Lys33smad3 and Ser37smad3 on the helix α2 are engaged in direct DNA phosphate backbone interaction in Smad3 while these contacts are disrupted in both Smad1 and Smad5 (Figure 2C). Previous work on Smad1 has shown that the structural differences of Smad1 with an ‘open’ and Smad3 with a ‘closed’ N-terminus could influence their potential to homodimerize on composite sites (28). With the open domain-swapped conformation, Smad5 also exhibits a strongly cooperative binding to the palindromic SBE DNA sequence, while Smad3 binds additively (Figure 1A). The four determined MH1-SBE complex structures exhibit nearly identical protein-DNA interaction mode, although helices α1 of these MH1 domains adopt different conformations. The TGF-β responsive R-Smad, Smad3, and the Co-Smad, Smad4, are both in the ‘closed’ state, while the two BMP-responsive R-Smads, Smad1 and Smad5, have the ‘open’ conformation. The MH1 domains of Smad1 and Smad5 also exhibited similar cooperative bindings to the palindromic SBE sequence in the gel shift assays. However, it is noteworthy that Smad1 and Smad5 possess inherent specificities and are not always interchangeable in vivo (41,42). As the DNA binding modes of their MH1 domains are very similar, the selectivity of Smad1 and Smad5 appears not to be achieved by the MH1 domain itself but instead through other mechanisms, such as the different transcriptional regulations for the two Smad genes (43).
Complex structure of Smad5-MH1 with GC-BRE DNA
In addition to the well characterized SBE sites, BMP-responsive Smad proteins are also often targeted to the GC-rich sites (8). The 6-bp palindromic sequence 5′-GGCGCC-3′ is widely accepted as a binding sequence for BMP-responsive Smads (18,23,44). As shown in Figure 1B, the Smad5 MH1 molecules bind more cooperatively to GC-BRE DNA than to the SBE DNA, suggesting a distinct mode for Smad5 to bind to the GC-rich sequences. To unravel the mechanism of Smad-MH1 binding to the GC-rich site, crystallization of the Smad5 MH1 domains complexed with the GC-BRE sequence was performed and the best sample diffracted to 3.05 Å (Table 1). The 14-bp DNA duplex in crystallization trials includes a central palindromic 5′-GGCGCC-3′ sequence flanked by 4 base pairs on each end (Figure 3A). The asymmetric unit contains two protein-DNA complexes with each containing two Smad5 MH1 monomers and a double-stranded DNA (Supplementary Figure S3A). These two 2:1 protein-DNA complexes are nearly identical with a Cα RMSD of 0.25 Å for 212 aligning residues. Within the Smad5-MH1/GC-BRE complex, two MH1 domains are located approximately on the same sides of the DNA and both proteins bind to the same major groove of the palindromic GC-BRE site (Figure 3A). The two MH1 monomers adopt essentially an identical conformation with an RMSD of 0.23 Å over 98 Cα atoms. When the N-terminal portion of Smad5 was modeled, the electron density map suggested that all of the four Smad5 monomers adopted the ‘closed’ conformation, in which helix α1 folds back to pack against helices α2 and α3 (Supplementary Figure S3B). Therefore, both the domain-swapped arrangement and the fold-back conformation of Smad5-MH1 were captured during our crystallization, in the SBE complexed structure and the GC-BRE complexed structure, respectively. This indicates that the MH1 domain of Smad5 could change between these two conformations and that the alternative conformations might be characteristic of the BMP Smads.
Figure 3.
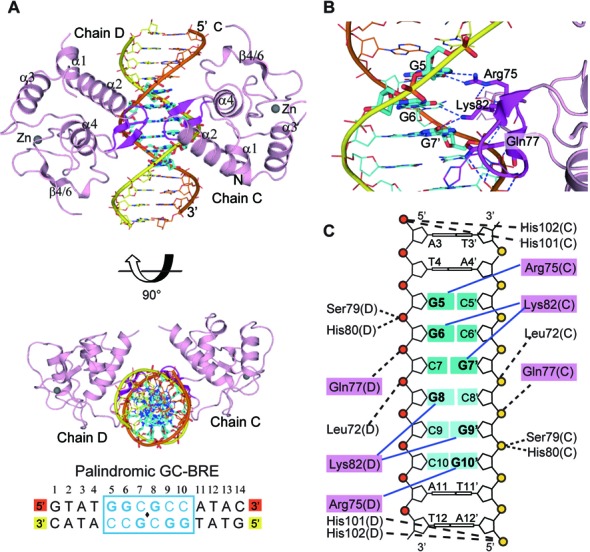
Crystal structure of Smad5-MH1 in complex with the GC-BRE sequence. (A) Overall structure of the Smad5-MH1/GC-BRE complex. The Smad5 MH1 domains are colored in light pink, with the β-hairpin highlighted in magenta. The zinc atoms are shown as spheres. The template chain and the complementary chain of the dsDNA are shown in orange and yellow, respectively. The DNA bases of the central GC-BRE site are shown as sticks and colored in cyan. The DNA sequence used for crystallization is shown below the structure, with the central 6-bp GC-BRE site boxed and highlighted in blue. (B) Detailed interactions between the β-hairpin and the DNA bases. Three conserved β-hairpin residues and the DNA bases involved in specific interactions are highlighted as magenta and cyan sticks, respectively. Hydrogen bonds are represented by blue dashed lines. (C) Schematic drawing of Smad5-MH1 binding to the GC-BRE sequence. Non-interacting nucleotides are omitted. Each copy of the GC-rich motif is shaded in dark cyan and light cyan, respectively. The conserved β-hairpin residues, Arg75, Lys82 and Gln77, are shaded in pink. Interactions between the amino acids and DNA bases are shown as blue solid lines, while the contacts with the DNA phosphates are shown as black dashed lines.
In the Smad5-MH1/GC-BRE complex, each of the two Smad5 MH1 domains recognizes half of the palindromic GC-BRE sequence (GGC) through the β-hairpin motif formed by β2 and β3 strands. The overall conformation of the β-hairpin is identical, but it binds to the two target sites (SBE and GC-rich) in different modes (Figure 3B, C and Supplementary Figure S3C). Arg75 and Lys82, two of the three invariant residues for SBE recognition, are still responsible for the base-specific contacts with the GC-BRE site. Arg75 forms two hydrogen bonds with the O6 and N7 of Gua5, which is the first base of the GC-BRE motif. Meanwhile, this bidentate interaction is reinforced by additional hydrogen bound from the carboxylate of Asp73 to the guanidinium group of Arg75, which has also been observed in Smad-SBE complexes. However, Lys82, which contacts the last two stacked bases in the SBE box, interacts with the diagonally-positioned DNA bases in the GC-BRE motif, Gua6 (the second position of the GGC motif) and Gua7’ (the complementary base of the third Cytosine in the GGC motif). These interactions are further stabilized by the moderate contact between the side chains of Lys82 and Gln77. Though Gln77 contacts the fourth base of the SBE box, it is not involved in the base-specific interaction in GC-BRE recognition. In addition to the base-specific interactions, the β-hairpin also forms several DNA backbone contacts with the DNA, including the interactions mediated by amides of Leu72, Gln77, Ser79 and His80 with the phosphate groups of Gua7’, Cyt8’ and Gua9’, respectively (Figure 3B and C). In addition to the recognition β-hairpin, two histidine residues (His101 and His102), located N-terminal to the β4 stand, are in close proximity to the backbone phosphate of the DNA, and the two imidazole rings approach the DNA like a clamp (Supplementary Figure S3D). The two histidine residues are highly conserved across R-Smads and the His-clamp is located closer to the DNA than all other R-Smad/palindromic SBE complex structures (27–29,40), which might contribute to the reinforcement of the protein-DNA contacts. The protrusion of helix α1 into the ‘open’ conformation of Smad5 and Smad1 disrupts several DNA contacts mediated by helix α2 in Smad3. However, no direct protein-DNA interaction in helix α2 is seen in the Smad5-MH1/GC-BRE complex, despite the N-terminal ‘closed’ conformation of the Smad5 MH1 monomer. In summary, our structure demonstrates that the 5′-GGC-3′ sequence represents the minimal GC-rich site recognized by the conserved β-hairpin of Smad5 and this interaction is strengthened by multiple backbone contacts.
The distinct structural characteristics of the Smad5-MH1/GC-BRE complex
In the 2:1 Smad5-MH1/GC-BRE complex, we found that the two MH1 molecules engaged in direct protein-protein interaction with each other. Although the Smad5 MH1 domains adopt similar conformation for assembly on the palindromic 5′-GGCGCC-3′ sequence as that on the palindromic SBE sequence (5′-GTCTAGAC-3′), the compressed 6-bp GC-BRE site leads to a closer location of the two MH1 domains on the DNA. The closest Cα-Cα distance between the two DNA bound MH1 molecules is only 5Å in the Smad5-MH1/GC-BRE complex (Figure 4A and B), while it is over 11Å in all of the Smad-MH1/SBE structures (Figure 4A and C). Therefore, in the Smad5-MH1/GC-BRE complex, the imidazole ring of His80 of one MH1 domain is able to contact the carbonyl oxygen of His80 in the other one (Figure 4B). This Histidine residue located in the loop connecting the β2 and β3 strands and this Ser-His dipeptide at the tip of the DNA recognition β-hairpin are highly conserved in all R-Smads (Figure 1D). Mutation of the histidine residue in Smad5 resulted in a decreased binding affinity with GC-BRE DNA, but the highly cooperative DNA binding mode is retained (Figure 4D). However, the Smad5 H80A mutation had no effect on the binding to palindromic SBE DNA (Supplementary Figure S4A). Therefore, the direct intermolecular interaction observed in the structure occurs while two MH1 molecules locate into the short 6-bp GC-BRE site and these physical contacts further stabilize the complex. Additionally, the N-terminus of helix α2 of one MH1 domain is also in close proximity to the N-terminus of helix α2 of the other one in the 2:1 MH1-DNA complex (Figure 4A). Although the linker between α2 and α1 was not modeled due to poor electron density, the trend of the peptide chain strongly suggests potential contacts between the two MH1 molecules. Collectively, direct protein-protein interactions occur when two MH1 monomers bind to the adjacent GC-rich sites which are more compact than SBE repeats. These intermolecular interactions may not be the driving force of the cooperative binding mode, but do make a contribution to the stability of the hetero-oligomeric complex.
Figure 4.
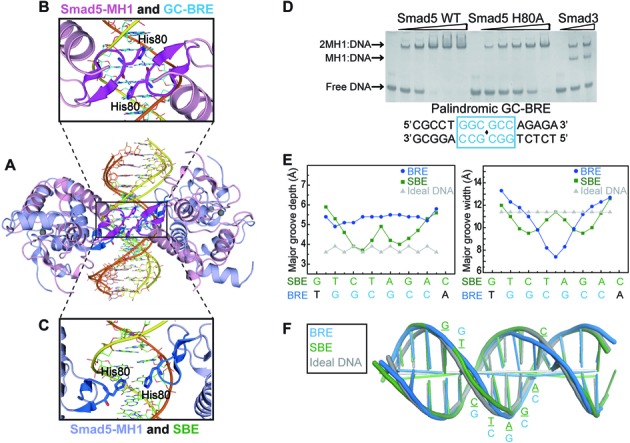
Structural comparisons between the Smad5-MH1/GC-BRE complex and the Smad5-MH1/SBE complex. (A) Structural superposition of the Smad5-MH1/GC-BRE complex with the Smad5-MH1/SBE complex. The color codes for the Smad5-MH1/SBE complex and the Smad5-MH1/GC-BRE complex follow that in Figures 2A and 3A, respectively. (B and C) Magnification of the region boxed in Figure 4A, displaying the relative position of two His80 residues from the two co-binding MH1 molecules in (B) the Smad5-MH1/GC-BRE complex and (C) the Smad5-MH1/SBE complex. (D) EMSA of Smad5-MH1 wild type (left panel of the gel) and H80A mutant (middle panel of the gel) bound to 1 μM of GC-BRE DNA. The protein concentrations used were 0, 1.25, 2.5, 3.75, 5, 6.25 μM (from left to right in each panel). The Smad3-MH1/SBE complex is shown as a molecular weight marker at the right panel of the gel. (E) Major groove depth and width are calculated using Curves+ for the Smad5-MH1 bound SBE DNA (green squares) and GC-BRE DNA (blue circles). Parameters for ideal B DNA are shown as grey triangles. (F) Overlay of the helical axes calculated with Curves+. Cartoons of the Smad5-MH1 bound SBE DNA (green), GC-BRE DNA (blue) and the ideal B DNA (grey) illustrate the differences within the overall curvature of the double helix.
The affinity, cooperativity and specificity of protein-DNA interaction can also be largely affected by the shape of DNA (45,46). Therefore, we analyzed the topology parameters of the Smad5-MH1 bound GC-BRE DNA using Curves+(36) and compared it with the Smad bound SBE DNA. The shape of the palindromic SBE DNA bound with Smad5 are identical to those bound with other R-Smad or the Co-Smad (Supplementary Figure S2C), and the Smad5 bound SBE DNA is mainly used in the following comparisons. The DNA duplex in the Smad5-MH1/GC-BRE complex is not involved in the crystal-packing interactions as one DNA duplex end is vertically packed against the other from the symmetry-related molecule, resulting in a 90° twist. The GC-BRE DNA could be largely characteristic for a standard B-form DNA with the exception of the strong compression of the major groove within the central palindromic binding site (Figure 4E and F). The width of the major groove at the center of the 2-fold axis (GGC·GCC) is significantly decreased, which is much more pronounced than the Smad5 bound SBE DNA. However, the Smad5 bound GC-BRE DNA exhibits the lowest overall bend (5.3°) when compared with all the R-Smad bound SBE DNA (15.6° for Smad5, 14.5° for Smad1 and 19.8° for Smad3) (Figure 4F). Other helical parameters of Smad5 bound GC-BRE DNA, such as roll, twist, slide and shift, also exhibit differences from the Smad5-bound SBE DNA (Supplementary Figure S4B-S4E). For example, at the palindromic center, neither the roll angle nor the twist angle is sharply changed in the GC-BRE DNA, while the corresponding angles in SBE DNA undergo pronounced deviations. Therefore, two MH1 monomers squeeze into the same major groove of the GC-BRE binding site and lead to a largely compressed major groove, indicating that these conformational changes could affect the cooperativity of Smad5 binding to different DNA sequences.
Structure of the Smad5 MH1 domains bound with composite Smad elements
In BMP signaling, the composite Smad binding sequences which contain both the GC-rich and SBE sites have been identified in many target genes and are often required for full BMP responsiveness (23). As shown in Supplementary Table S1, these composite sequences contain a 6-bp GC-rich element, flanked by one or multiple SBE sites. To unravel the molecular mechanism of Smad-MH1 recognizing adjacent GC-rich and SBE sites, crystallization trials of Smad5-MH1 in complex with several native composite DNAs were carried out, but no crystal appeared (Supplementary Table S2). We therefore modified the DNA sequence by adding SBE site to both ends of the palindromic GC-rich elements (5′-GGCGCC-3′or 5′-GCCGGC-3′) and/or varying the spacer length between the GC-rich and SBE sites. The Smad5-MH1 protein binds to all these native and modified composite sequences in a highly cooperative manner (Figure 1C, Supplementary Figures S1, S5A and S5B). After extensive attempts, a DNA duplex (GCRj2) comprising a central palindromic GCCGGC sequence flanking with a 4-bp single SBE element on both ends produced good diffracting crystals to 3.2 Å resolution (Figure 5A and Table 1). Although the data set is at medium resolution, the molecular replacement solution was robust and yielded an initial electron density that was good enough to reveal the location of DNA phosphate backbones and base stacks, as well as the MH1 domains. All 22 nucleotides of the GCRj2 DNA were identified in the model, and the dsDNA molecules form pseudo-continuous duplexes in which the thymine (T) overhang of one duplex pairs with the adenine (A) nucleotide overhang of another symmetric molecule. There are four Smad5 MH1 domains and two duplexes of DNA molecules in an asymmetric unit (Supplementary Figure S5C). Each two MH1 domains bind to one GCRj2 DNA, forming a 2:1 protein-DNA complex, and each of the sequentially arranged MH1 domains on one DNA duplex adopts a similar ‘open’ conformation and protrudes its helix α1 to contact the hydrophobic interface formed by helix α2 and α3 of the MH1 monomer on another DNA duplex, generating an integral 4:2 protein-DNA complex in the asymmetric unit. The two 2:1 protein-DNA complexes in one asymmetric unit are almost identical with a Cα RMSD of 0.51 Å for the overall 217 atoms. Within the GCRj2 DNA sequence, there are four potential MH1 binding sites, the central two GC sites (GCC and GGC) and two flanking 4-bp SBE boxes (AGAC and GTCT) (Figure 5A). The EMSA assay demonstrates that the assembly of Smad5-MH1 on the GCRj2 DNA exhibits a significantly cooperative binding pattern and forms an oligomeric complex (Supplementary Figure S5B). However, in the crystal structure, only two MH1 monomers are located at two distant sites of the sequence, the first half of the GC-rich site (‘GCC’ site, Site 2) and the 3′-terminus single SBE box (‘GTCT’ site, Site 4) (Figure 5A and B). Were the open conformational Smad5-MH1 to be located at either Site 1 or Site 3, its helix α1 would severely clash with the symmetric molecule. Therefore, it is impossible for four MH1 molecules to be recruited to the consecutive binding sites in the crystal structure due to the crystal packing.
Figure 5.
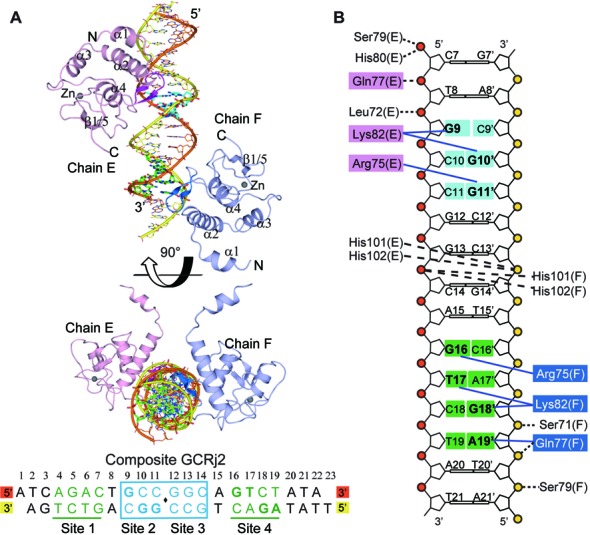
Crystal Structure of Smad5-MH1 in complex with a composite DNA element (GCRj2). (A) Overall structure of the Smad5-MH1/GCRj2 complex. The GCRj2 DNA sequence used for crystallization is shown at the bottom and potential binding sites are labeled. The Smad5 MH1 domains bound to the GC-rich site and the SBE site are shown as that in Figures 3A and 2A, respectively. The DNA bases of Site 2 (GC-rich site) and Site 4 (SBE site) are highlighted as sticks and colored in cyan and green, respectively. (B) Schematic representation of the interactions between the Smad5 MH1 domain and the GCRj2 sequence. Amino acids and DNA bases in the recognitions of Site 2 and Site 4 are shown as that in Figures 3C and 2C, respectively.
Similar to our other two MH1-DNA complex structures, bindings of Smad5-MH1 to Site 2 and Site 4 of the GCRj2 DNA are also mediated by the conserved β-hairpin (β2 and β3), which inserts into the major grooves of the DNA duplex of the target sites. The two binding sites are separated by four base pairs and are located on the opposite faces of the dsDNA. The 5′-GCC-3′ site is the reverse complement of the single copy of the minimal GC-rich site (5′-GGC-3′). Indeed, the Smad5 MH1 domain uses a similar protein-DNA interaction mode to recognize the 5′-GCC-3′ site in GCRj2 as it binds to the GC-BRE sequence. Arg75 interacts with O6 and N7 of Gua11’ while Lys82 makes base-specific contacts with the diagonally positioned Gua9 and Gua10’ (Figure 5B and Supplementary Figure S5D). In the single SBE site which was bound by Smad5, the interactions involve the same β-hairpin residues as those acting in recognition of the palindromic SBE sequence, but they formed slightly different amino acid-base contacts (Figure 5B and Supplementary Figure S5E). The conserved residues, Arg75 and Gln77, still make base-specific contacts with G16 and A19’, respectively. However, the rotation of the side-chain of Lys82 allows it to contact the diagonally positioned DNA bases, T17 and G18’ of the single SBE site, while failing to contact Gln77. Apart from the base-specific recognitions, the phosphate backbone contacts at two Smad binding sites are similar, mediated by the main-chain amides of Ser71, Leu72, Gln77 and Ser79, as well as the imidazole ring of His101 and His102. Therefore, together with structures of the Smad5-MH1-SBE complex and the Smad5-MH1-GC-BRE complex, the complex structure of Smad5-MH1 and the composite GCRj2 DNA demonstrates that the minimal binding sites for the Smad MH1 domain are 5′-GGC-3′ (GC-rich site) and 5′-GTCT-3′ (SBE site), and that the heterotrimeric Smad complex can recognize the composite sites through a modular binding mode.
DISCUSSION
Smad 1/5/8 are the mediators of BMP signaling, and direct binding of the proteins to the target sites is fundamental to signaling transduction. Both GC-rich and SBE sites are required for full BMP-responsive gene expressions, but previous research has only revealed how R-Smads and the Co-Smad recognize and directly bind to the SBE site (27–29). The 6-bp GC-rich sequence 5′-GGCGCC-3′ (GC-BRE) and its variant sequences are enriched in the cis-regulatory elements of multiple BMP-responsive genes (Supplementary Table S1) (18,20,21,23,44). The in vitro binding assay demonstrated that GC-BRE DNA is contacted by two MH1 domains of R-Smads (17). Through mutational analysis and homology modeling, it was proposed that recognition of the GC-rich site by MH1 employs all three β-hairpin contacting residues to bind to the SBE site (Arg75, Gln77 and Lys82, numbered in Smad5). Compared with the 8-bp palindromic SBE motif, the GC-rich site spans only 6 base pairs, therefore previous model suggested that two β-hairpins from each MH1 domain point toward each other and overlap across the two central base pairs (17). Here we report the first crystal structure of the Smad MH1 domain binding to the GC-rich DNA sequences, thus revealing the binding mode of Smad proteins to the different types of DNA sequences. Our structures demonstrate that the 3-bp site, 5′-GGC-3′, represents the minimal GC-rich binding motif for Smad, which can exist as a single copy or in a palindromic repeated form. The β-hairpin of the MH1 domain contacts the specific DNA bases at the major groove both in the GC-rich site and the SBE site, and the base-specific DNA bindings are enabled by direct hydrogen bond interactions involving different pairings of amino acids and bases. The rearranged amino acid-base contacts allow the accommodation of two MH1 domains at the same side of the compressed 6-bp GC-rich site. Moreover, the compact binding mode results in direct intermolecular contacts between the two MH1 domains, mediated by a histidine residue in the loop region of the β-hairpin (His80 in Smad5). This residue is highly conserved among R-Smads but is replaced by glycine in Smad4. It was reported that substitution to a bulky residue in place of this histidine in Mad (R-Smad homologue in Drosophila) caused a substantial reduction in the binding affinity for the target gene (17). Here we also show that the H80A mutant of the Smad5-MH1 exhibits impaired binding affinity but still allows cooperative binding to the GC-BRE sequence. However, Smad3 binds to the GC-BRE sequence in a less cooperative manner, despite its identical β-hairpin constitution with Smad5, while the Smad4 MH1 domain forms a rather unstable monomeric complex. Taking together, these data suggest that the histidine residue would assist in facilitating the binding of two MH1 domains to the symmetric GC-BRE sequence, but the driving force for the specific binding patterns adopted by each Smad protein is comprehensive.
Hydrogen bonds provide specificity in amino acid-base interaction in most protein-DNA complexes (45–47). While inspecting the base-specific contacts of Smad binding to the SBE (5′-GTCT-3′) or GC-rich sites (5′-GGC-3′), we found that Arg75 and Lys82 always make conserved residue-base contacts with the two guanines at position 1 and 3′ of the SBE or GC-rich motifs. Lys82 can be a multiple donor amino acid upon DNA binding. It makes hydrogen bonds with position 4′ and Gln77 in the palindromic SBE binding, or with position 2 in single SBE site. While binding to the GC-rich site, Lys82 contacts guanine at position 2 and the carbonyl group of Gln77. Moreover, Gln77 makes additional contact with the adenosine base at position 4′ in SBE but not in the GC-rich DNA recognition. Specificity conveyed through hydrogen bonds depends on the number of contacts formed between protein residues and DNA bases and also on the uniqueness of the hydrogen bonding geometry (45). Therefore, the different bidentated hydrogen bonds conducted by Lys82 and Gln77 imply a higher binding affinity and specificity for Smad-MH1 to the 5′-GTCT-3′ single site than to the 5′-GGC-3′ site. In addition, it was proposed that the DNA structure substantially affects protein–DNA binding through indirect readout mechanisms (45,46). We therefore analyzed the DNA parameters in the GC-rich sequence or the SBE sequence and found distinctive changes in the DNA shapes of different binding sequences. In the palindromic SBE site, the symmetric center is strongly overwound and has an increased depth of the minor groove, whereas in the center of the palindromic GC-BRE site, there is a remarkable reduction in the width of the major groove, which could stabilize the binding of the protein. Therefore, our structures demonstrated that although the DNA-recognition β-hairpin is conserved in all R-Smads and the Co-Smad, Smad-MH1 can use both the base readout and the local DNA shape readout mechanism to recognize the GC-rich and SBE DNA sequences. Additionally, Smad protein can bind to certain SBE variants (such as GGCTT or GACG) or diverse GC-rich elements (such as GRCGNC) found by genome-wide sequence analysis (21,48). For efficient binding, all of them would need to retain the crucial bidentate contacts between Arg75 and the guanine at position 1. The relaxed binding specificity is satisfied by the bifurcated bonds from Lys82 or Glu77, as the Lys82 residue has the ability to form interactions with the directly stacked or diagonally positioned DNA bases pairs. Thus, the interaction modes illuminated in our crystal structures shed light on the mechanism of both DNA binding specificity and degenerate bases tolerance.
Decapentaplegic (Dpp) is a well-characterized TGF-β/BMP family member in Drosophila. Previous studies have identified two short DNA motifs in Dpp signaling, SE and AE (silencer element and activating element, respectively). Both of these two motifs are bipartite elements consisting of a GC-rich site bound by Mad and a SBE site bound by Medea, and spaced by several nucleotides (4-bp for AE and 5-bp for SE, respectively) (38,49). The transcription response activity of SE strictly depends on the linker length, while some spacer flexibility is allowed in AE. However, the linker-length does not restrict the formation of the Mad-Medea complex in vitro in both cases. Such bipartite elements have also been found in many BMP-responsive genes in other species (Supplementary Table S1). To investigate the binding mode of Smad on native bipartite elements, we modeled the complex of three MH1 monomers binding with a sequence from Id2, based on the structures of Smad-MH1 in complex with either the SBE or GC sequences (Figure 6A). Two Smad MH1 domains contact the GC-BRE sequence while one Smad MH1 monomer binds with the single SBE site which is separated by a 5-bp spacer from the GC-BRE site. There is sufficient space for the Smad C-terminal MH2 domains to become involved. In addition, we changed the length of the spacer in the Id2 sequence and examined the assembly of the Smad5 MH1 domains on these DNA mutants (Supplementary Figure S6). At a represented concentration, the Smad5 MH1 molecules cooperatively bind with the Id2 DNA and they form a 3:1 protein-DNA complex. Interestingly, the maximum binding occurs at the 3–4 bps spaced DNA mutants while the affinity to the 2-bp spaced one remains higher than that of the native DNA (Figure 6B). Moreover, DNA mutants with a 6-bp linker or no spacer exhibited a dramatically unstable binding to the Smad5 MH1 domain, with the complex migrating as poorly resolved bands in the gel (Supplementary Figure S6D). Even though the linker variation does not significantly influence the assembly of the three MH1 domains on the composite binding site, it would greatly change the location pattern of the heterotrimeric Smad complex on the DNA sequence. Because precise distances and hydrogen bond numbers have great significance in energy function, these apparently similar interactions actually give rise to distinct energetic patterns for individual Smad proteins to the DNA. Numerous studies have suggested that Smad transcription complexes often colocalize with other master transcription factors that specify and maintain the cell identities (50–53). Therefore, we suggest that the spacer region in the bipartite element together with the cooperative actions of Smad proteins with their cofactors may influence the assembly of the different transcription complexes on target sequences, which can lead to diverse gene regulations and phenotype changes.
Figure 6.
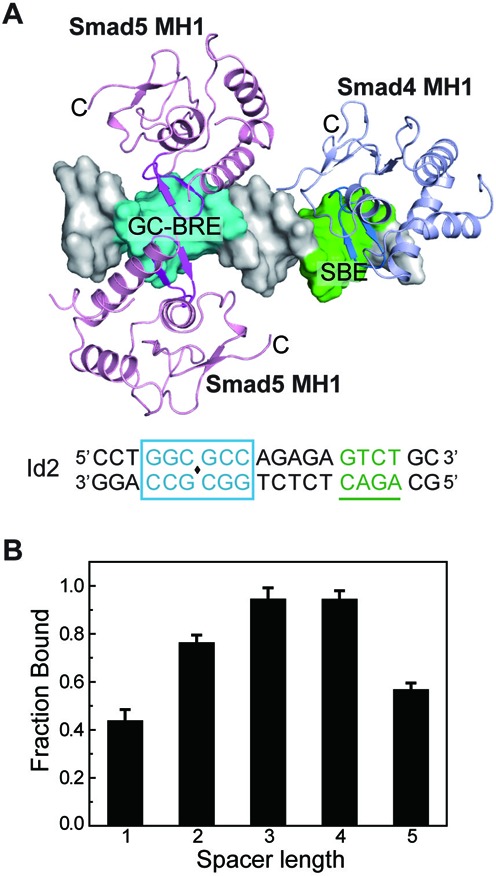
Analysis of the binding of Smad5-MH1 to the composite DNA elements. (A) Modeled structure of the Smad MH1 domains in complex with a DNA sequence in Id2 promoter. The DNA sequence is shown at the bottom as that in Figure 1C. Two molecules of Smad5-MH1 (derived from Smad5-MH1/GC-BRE complex structure) and one molecule of Smad4 MH1 domain (derived from PDB id: 3QSV) are modeled to the Id2 promoter DNA. The DNA duplex is shown as a white surface with the GC-rich site and SBE site colored in cyan and green, respectively. The MH1 domains of Smad4 and Smad5 are colored in light pink and light blue, respectively. The C-terminals of each MH1 protein are labeled. (B) Assembly of Smad5-MH1 on composite DNA elements with different linker lengths. EMSA assays were performed in the presence of 2.1 μM Smad5-MH1 protein and 1 μM DNA with different linker lengths between the bipartite elements. Error bars are plotted as ±SEM from at least three independent experiments. Representative EMSA assays of the Smad5 MH1 proteins binding to these DNA variants are shown in Supplementary Figure S6.
ACCESSION NUMBERS
Coordinates and structure factors have been submitted to the protein databank with the accession numbers 4ZKG, 4ZL2 and 4ZL3.
Supplementary Material
Acknowledgments
We thank J.H. He, L.F. Yang and other staff members of beamline BL17U at SSRF for assistance in data collection. We also appreciate the help from C.Y. Yan in structure determination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31130062 and 31321003 to J.W.W.]; China National Basic Research Program [2013CB530600 to J.W.W. and 2012CB967900 to S.M.Y.]. Funding for open access charge: National Natural Science Foundation of China [31130062].
Conflict of interest statement. None declared.
REFERENCES
- 1.Massagué J., Blain S.W., Lo R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu M.Y., Hill C.S. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev. Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Groppe J., Hinck C.S., Samavarchi-Tehrani P., Zubieta C., Schuermann J.P., Taylor A.B., Schwarz P.M., Wrana J.L., Hinck A.P. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 4.Kang J.S., Liu C., Derynck R. New regulatory mechanisms of TGF-beta receptor function. Trends Cell Biol. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Massagué J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 6.Massagué J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 7.Heldin C.H., Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng X.H., Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 10.Wrana J.L., Attisano L., Wieser R., Ventura F., Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 12.Zawel L., Dai J.L., Buckhaults P., Zhou S., Kinzler K.W., Vogelstein B., Kern S.E. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 13.Dennler S., Itoh S., Vivien D., Dijke P., Huet S., Gauthier J.M. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denissova N.G., Pouponnot C., Long J., He D., Liu F. Transforming growth factor beta -inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6397–6402. doi: 10.1073/pnas.090099297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X., Yang J., Li Y., Liu Y. Both Sp1 and Smad participate in mediating TGF-beta1-induced HGF receptor expression in renal epithelial cells. Am. J. Physiol. Renal. Physiol. 2005;288:F16–F26. doi: 10.1152/ajprenal.00318.2003. [DOI] [PubMed] [Google Scholar]
- 16.Jonk L.J., Itoh S., Heldin C.H., ten Dijke P., Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 17.Gao S., Laughon A. Decapentaplegic-responsive silencers contain overlapping mad-binding sites. J. Biol. Chem. 2006;281:25781–25790. doi: 10.1074/jbc.M603371200. [DOI] [PubMed] [Google Scholar]
- 18.Karaulanov E., Knöchel W., Niehrs C. Transcriptional regulation of BMP4 synexpression in transgenic Xenopus. EMBO J. 2004;23:844–856. doi: 10.1038/sj.emboj.7600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida W., Hamamoto T., Kusanagi K., Yagi K., Kawabata M., Takehara K., Sampath T.K., Kato M., Miyazono K. Smad6 is a Smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J. Biol. Chem. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 20.López-Rovira T., Chalaux E., Massagué J., Rosa J.L., Ventura F. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J. Biol. Chem. 2002;277:3176–3185. doi: 10.1074/jbc.M106826200. [DOI] [PubMed] [Google Scholar]
- 21.Korchynskyi O., ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 22.Kim J., Johnson K., Chen H.J., Carroll S., Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa M., Koinuma D., Tsutsumi S., Vasilaki E., Kanki Y., Heldin C.H., Aburatani H., Miyazono K. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 2011;39:8712–8727. doi: 10.1093/nar/gkr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Candia A.F., Watabe T., Hawley S.H., Onichtchouk D., Zhang Y., Derynck R., Niehrs C., Cho K.W. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 25.Xiao L., Yuan X., Sharkis S.J. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 26.Grönroos E., Kingston I.J., Ramachandran A., Randall R.A., Vizan P., Hill C.S. Transforming growth factor beta inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol. Cell. Biol. 2012;32:2904–2916. doi: 10.1128/MCB.00231-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y., Wang Y.F., Jayaraman L., Yang H., Massagué J., Pavletich N.P. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 28.BabuRajendran N., Palasingam P., Narasimhan K., Sun W., Prabhakar S., Jauch R., Kolatkar P.R. Structure of Smad1 MH1/DNA complex reveals distinctive rearrangements of BMP and TGF-beta effectors. Nucleic Acids Res. 2010;38:3477–3488. doi: 10.1093/nar/gkq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baburajendran N., Jauch R., Tan C.Y., Narasimhan K., Kolatkar P.R. Structural basis for the cooperative DNA recognition by Smad4 MH1 dimers. Nucleic Acids Res. 2011;39:8213–8222. doi: 10.1093/nar/gkr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makkar P., Metpally R.P., Sangadala S., Reddy B.V. Modeling and analysis of MH1 domain of Smads and their interaction with promoter DNA sequence motif. J. Mol. Graph. Model. 2009;27:803–812. doi: 10.1016/j.jmgm.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchet C., Pasi M., Zakrzewska K., Lavery R. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011;39:W68–W73. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyrowolakis G., Hartmann B., Müller B., Basler K., Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev. Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Weiss A., Charbonnier E., Ellertsdóttir E., Tsirigos A., Wolf C., Schuh R., Pyrowolakis G., Affolter M. A conserved activation element in BMP signaling during Drosophila development. Nat. Struct. Mol. Biol. 2010;17:69–76. doi: 10.1038/nsmb.1715. [DOI] [PubMed] [Google Scholar]
- 39.Benchabane H., Wrana J.L. GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol. Cell. Biol. 2003;23:6646–6661. doi: 10.1128/MCB.23.18.6646-6661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chai J., Wu J.W., Yan N., Massagué J., Pavletich N.P., Shi Y. Features of a Smad3 MH1-DNA complex. Roles of water and zinc in DNA binding. J. Biol. Chem. 2003;278:20327–20331. doi: 10.1074/jbc.C300134200. [DOI] [PubMed] [Google Scholar]
- 41.McReynolds L.J., Gupta S., Figueroa M.E., Mullins M.C., Evans T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110:3881–3890. doi: 10.1182/blood-2007-04-085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dick A., Meier A., Hammerschmidt M. Smad1 and Smad5 have distinct roles during dorsoventral patterning of the zebrafish embryo. Dev. Dyn. 1999;216:285–298. doi: 10.1002/(SICI)1097-0177(199911)216:3<285::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 43.Wei C.Y., Wang H.P., Zhu Z.Y., Sun Y.H. Transcriptional factors smad1 and smad9 act redundantly to mediate zebrafish ventral specification downstream of smad5. J. Biol. Chem. 2014;289:6604–6618. doi: 10.1074/jbc.M114.549758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakahiro T., Kurooka H., Mori K., Sano K., Yokota Y. Identification of BMP-responsive elements in the mouse Id2 gene. Biochem. Biophys. Res. Commun. 2010;399:416–421. doi: 10.1016/j.bbrc.2010.07.090. [DOI] [PubMed] [Google Scholar]
- 45.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frederick J.P., Liberati N.T., Waddell D.S., Shi Y., Wang X.F. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao L.C., Blitz I.L., Peiffer D.A., Phin S., Wang Y., Ogata S., Cho K.W., Arora K., Warrior R. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development. 2006;133:4025–4034. doi: 10.1242/dev.02561. [DOI] [PubMed] [Google Scholar]
- 50.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 51.Trompouki E., Bowman T.V., Lawton L.N., Fan Z.P., Wu D.C., DiBiase A., Martin C.S., Cech J.N., Sessa A.K., Leblanc J.L., et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mullen A.C., Orlando D.A., Newman J.J., Lovén J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isogaya K., Koinuma D., Tsutsumi S., Saito R.A., Miyazawa K., Aburatani H., Miyazono K. A Smad3 and TTF-1/NKX2–1 complex regulates Smad4-independent gene expression. Cell Res. 2014;24:994–1008. doi: 10.1038/cr.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


