Figure 4.
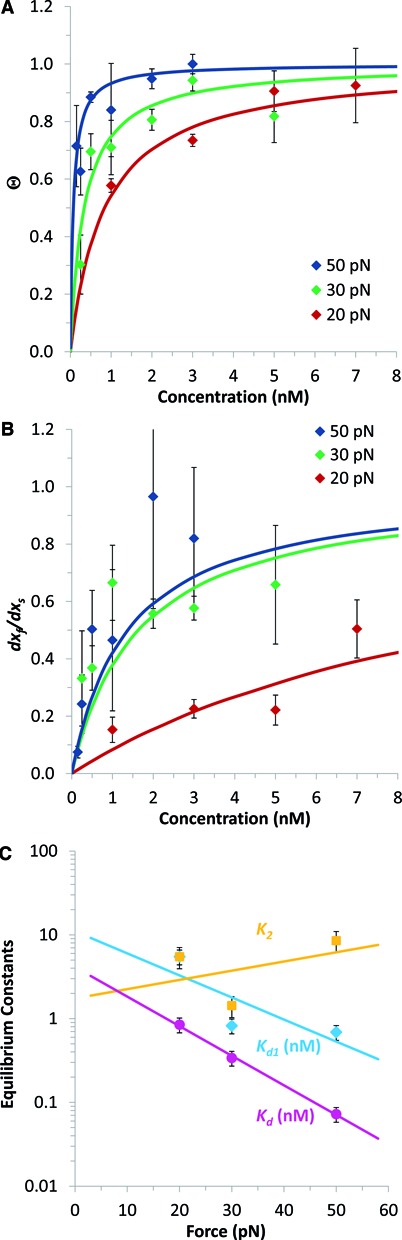
Equilibrium analysis of DNA elongations induced by flex-Ru2 intercalation. (A) Measured equilibrium flex-Ru2/DNA length expressed as an occupancy (Θ, relative to the saturated values of Figure 2) as a function of ligand concentration (C) for the forces of 20, 30 and 50 pN (red, green and blue). Fits to Equation (12) (lines) determine the binding constant Kd for each force. (B) The ratio of the fast and slow elongation amplitudes, dxf/dxs, as a function of C (20 pN: red, 30 pN: green and 50 pN: blue). Fits of these data points to Equation (15) (lines) determine Kd1(F). (C) Binding constants Kd1 (cyan), Kd (magenta) and K2 (gold) versus force, as obtained from the fits of the data in Figure 4A and B to Equation (10) (lines), with K2 calculated as K2 = Kd1/Kd. Fitted zero-force binding constants and the flex-Ru2/DNA length changes associated with each K are collected in Table 2.
