Figure 4.
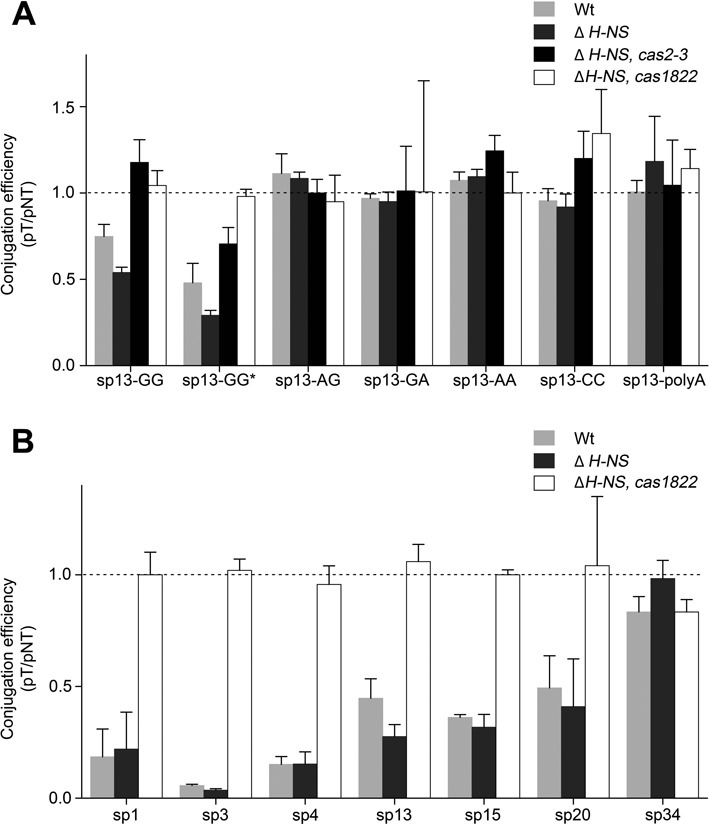
Analysis of the DNA target specificity of the S. putrefaciens CN-32 CRISPR–Cas system. Conjugation assays (see Figure 3) were used to analyze plasmid DNA target variants for DNA interference in S. putrefaciens CN-32 cells. (A) Interference was observed if the transferred DNA strand contained a sequence complementary to the crRNA13 (sp13-GG) or a sequence identical with spacer13 (sp13-GG*). Interference was evident for the wild-type and ΔH-NS strains, which contained all cas genes. However, the deletion of cas3 (Sputcn32_1820) or cas1822 abolished interference activity. The presence of the PAM sequence GG at the 3′ end of the DNA target (5′-protospacer-PAM-3′) was essential for the interference activity and PAM mutants (sp13-AG, sp13-GA, sp13-AA or sp13-CC) and a 10 nt poly-A sequence disrupting crRNA/DNA complementarity (sp13-polyA) were not targeted. (B) DNA interference activity was dependent on spacer sequence or crRNA abundance (spacers 1,3,4,13,15,20,34, see Figure 2).
