Figure 1.
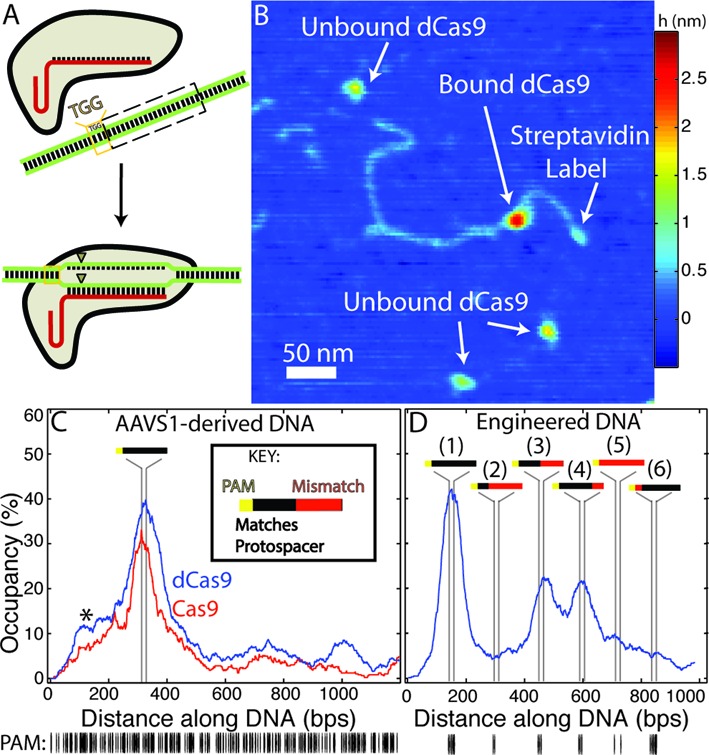
(A) Schematic representation of Cas9 activity: having bound a single-guide RNA (sgRNA, red), the Cas9-sgRNA complex binds to 20 bp ‘protospacer’ sequences in a targeted DNA molecule, provided that the protospacer is directly followed by a protospacer adjacent motif (PAM, here ‘TGG’). Following binding, the Cas9 endonuclease produces double-strand breaks (triangles) within the protospacer. (B) Atomic force microscopy (AFM) image of dCas9-sgRNA bound at the protospacer sequence within a single streptavidin-labelled DNA molecule derived from the human AAVS1 locus. (C–D) Fraction of bound DNA occupied by Cas9/dCas9-sgRNA along an AAVS1-derived (C) or an engineered DNA substrate (D) designed with a series of fully-complementary and partially-complementary protospacer sequences. Vertical lines represent the (23 bp) segments where each significant feature is located on the respective substrates (see inset key). (C) dCas9 and Cas9 exhibit nearly identical binding distributions on the AAVS1 substrate (n = 404 and n = 250, respectively). The asterisk marks an off-target ‘shoulder peak’ in the binding distribution (see text). (D) On the engineered substrate (n = 536) dCas9 binds with the highest propensity to the complete protospacer with no mismatched (MM) sites (peak 1, later referred to as the full or ‘0MM’ site) and also to sites with 10 or 5 mismatched bases distal to the PAM site (third and fourth feature from streptavidin label, referred to later as the ‘10MM’ or ‘5MM’ sites, respectively) albeit with the reduced affinity. Sites containing greater numbers of mismatches (second and fifth feature), or which possess two PAM-proximal mismatched nucleotides (sixth feature) are bound at significantly lower rates. (below) Distribution of PAM (‘NGG’) sites in each substrate.
