Figure 1.
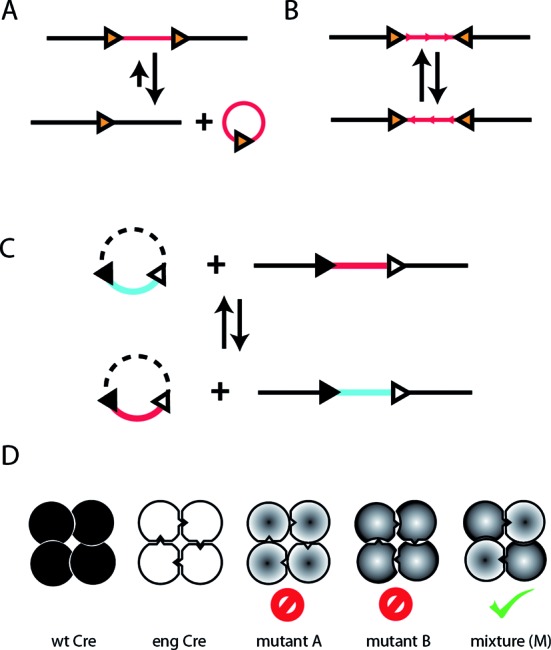
Genomic applications of Cre recombinase. Depending on the number and relative orientation of the loxP sites, Cre recombinase can perform deletion, inversion, insertion or exchange of genetic content. (A) Direct repeats of the loxP site can be recombined to excise the intervening genetic interval (downward arrow). This reaction is also catalyzed in the reverse direction, yielding a genetic insertion (upward arrow). For thermodynamic reasons, the excision reaction is favored and insertion events occur with low frequency. (B) Inverted loxP repeats can be recombined to yield an inversion of the bracketed DNA. (C) Recombination at pairs of distinct RT sites gives rise to exchange of the intervening genetic ‘cassette’. (D) Cre recombinase is a homotetramer in its functional complex (wt Cre), imparting a preference for a symmetric RT as a consequence. As a first step to achieving recombination at asymmetric sites, we desire an orthogonal engineered interface between Cre monomers (eng Cre). We seek to construct a novel homotetramer Cre mutant with monomer–monomer interfaces that, while functional, are incompatible with the wild-type protein. Combining wild-type and engineered half-interfaces gives rise to two distinct mutants that cannot form functional complexes (mutants A and B). Combining the two mutants (denoted by ‘M’) can reconstitute a functional heterotetrameric complex, which contains two wild-type and two engineered interfaces.
