Figure 3.
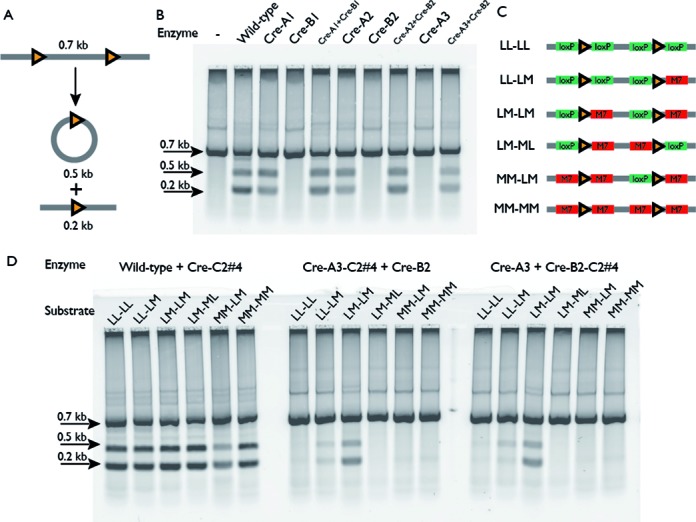
In vitro recombination assay of Cre mutants. (A) In vitro recombinase assay. A 0.7 kb linear DNA substrate with direct repeats of the loxP site (orange triangles) is incubated with wild-type or mutant Cre recombinase. The activity of functional Cre complexes results in production of a 0.5 kb circular product and a 0.2 kb linear product through intra-molecular excision. (B) In vitro assay results. Lane 1: DNA substrate alone; lane 2: wild-type Cre; lanes 3–5: first round redesigned Cre mutants Cre–A1, Cre–B1 and a mixture of the two (CreA1 + CreB1); lanes 6–8: second round redesigned Cre mutants Cre–A2, Cre–B2 and a mixture of the two (Cre–A2 + Cre–B2); lanes 9–10: third round mutant Cre–A3 and a mixture of Cre–A3 + Cre–B2. All Cre–B mutants are inactive in isolation. Cre–A mutants progressively lose homotetramer activity through the three rounds of design. (C) In vitro substrates for asymmetric recombination target site experiments. RT half-sites in the linear DNA substrate described in panel (A) were systematically varied to incorporate the M7 sequence (13). LoxP and M7 half-sites are rendered as green and red boxes, and abbreviated by the letters L and M, respectively. Combinations ranged from entirely loxP (LL–LL, the same as in panel (A)) to entirely M7 (MM–MM), including hybrid RT sites situated as both direct (LM–LM) and inverted (LM–ML) repeats. (D) The effect of controlled assembly of heterotetrameric Cre complexes. Each of the mixed loxP/M7 substrates was incubated with a pair of recombinases, one with mutations that recognize the M7 RT halfsite (Cre–C2#4) and the other with preference for the loxP half-site. In the left panel, the two proteins have no additional mutations to control complex formation. In the middle and right panels, recombinases with different RT specificities are combined with the Cre–A3 and Cre–B2 mutations, with both possible combinations tested. The restriction of permissible substrates by the Cre–A3 and Cre–B2 mutations are consistent with a requirement for an (ABAB) heterotetramer to achieve recombinase activity.
