Figure 4.
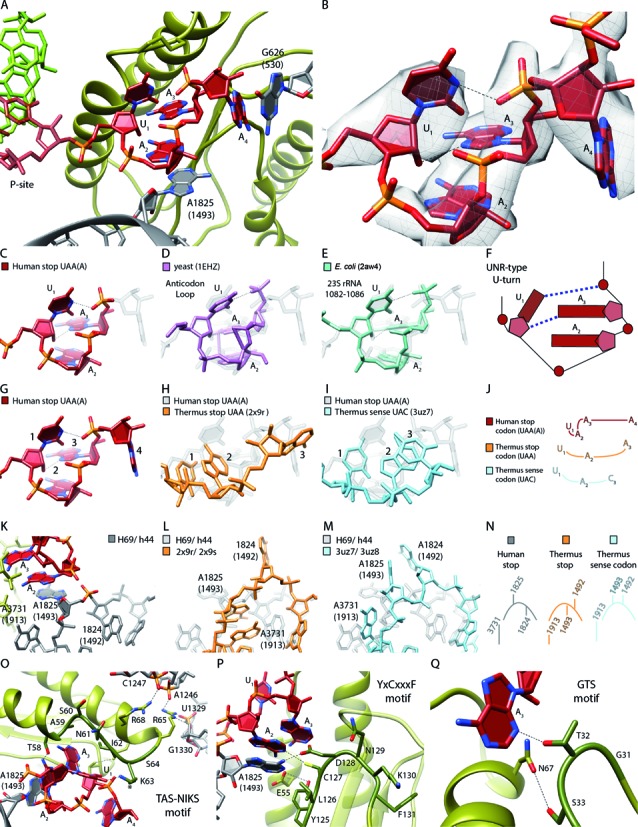
U-turn-like geometry of the UAA stop-codon bound by eRF1 and the ribosome. (A) Human UAA(A) stop-codon (red) positioned in a cavity between eRF1 (green) and the ribosomal bases Hs G626 (Ec G530) and Hs A1825 (Ec A1493) (gray). (B) EM density revealing the geometry of the mRNA sequence UAA(A) during stop codon recognition. (C–F) Comparison of the human stop codon mRNA geometry with the known UNR-type U-turn structures (D) yeast anticodon loop (purple) and (E) Ec 23S rRNA (1082–1086) (turquois). (F) Schematic representation of (C–E). (G–J) Comparison of the A-site mRNA geometry during (G) human UAA stop codon recognition (H), Thermus thermophilus (Tt) UAA stop codon recognition (light orange) and (I) Tt UAC sense decoding (light blue). (J) Schematic representation of (G–I). (K–N) Comparison of the h44 (Hs A1824 (Ec A1492), Hs A1825 (Ec A1493))/H69 (Hs A3731 (Ec A1913)) states during (K) human UAA stop codon recognition, (L) Tt UAA stop codon recognition and (M) Tt UAC decoding. (N) Schematic representation of (K–M). (O) Interactions of the eRF1 TAS-NIKS motif (residues 58–64) with the mRNA/rRNA. The hydroxylation site of K63 is located at C4 (*). (P) Interactions of the eRF1 YxCxxxF (residues 125–131) motif with the mRNA/rRNA. (Q) Interactions of the eRF1 GTS (residues 31–33) motif with the mRNA.
