Abstract
The eukaryotic microsomal cytochrome P450 systems consist of a cytochrome P450 enzyme (P450) and a cytochrome P450 redox partner, which generally is a cytochrome P450 reductase (CPR) that supplies electrons from NADPH. However, alternative electron donors may exist such as cytochrome b5 reductase and cytochrome b5 (CBR and CYB5, respectively) via, which is NADH-dependent and are also anchored to the endoplasmic reticulum. In the carotenogenic yeast Xanthophyllomyces dendrorhous, three P450-encoding genes have been described: crtS is involved in carotenogenesis and the CYP51 and CYP61 genes are both implicated in ergosterol biosynthesis. This yeast has a single CPR (encoded by the crtR gene), and a crtR - mutant does not produce astaxanthin. Considering that this mutant is viable, the existence of alternative cytochrome P450 electron donors like CBR and CYB5 could operate in this yeast. The aim of this work was to characterize the X. dendrorhous CBR encoding gene and to study its involvement in P450 reactions in ergosterol and carotenoid biosynthesis. Two CBRs genes were identified (CBR.1 and CBR.2), and deletion mutants were constructed. The two mutants and the wild-type strain showed similar sterol production, with ergosterol being the main sterol produced. The crtR - mutant strain produced a lower proportion of ergosterol than did the parental strain. These results indicate that even though one of the two CBR genes could be involved in ergosterol biosynthesis, crtR complements their absence in the cbr - mutant strains, at least for ergosterol production. The higher NADH-dependent cytochrome c reductase activity together with the higher transcript levels of CBR.1 and CYB5 in the crtR - mutant as well as the lower NADH-dependent activity in CBS-cbr.1- strongly suggest that CBR.1-CYB5 via participates as an alternative electron donor pathway for P450 enzymes involved in ergosterol biosynthesis in X. dendrorhous.
Introduction
The cytochrome P450s (P450s) constitute a large superfamily of heme-containing monooxygenases present in organisms from all domains of life [1,2]. They play significant roles in the oxidative metabolism of a wide range of exogenous and endogenous substrates [3]. These enzymes are involved in the metabolism of physiologically important compounds such as sterols, fatty acids and vitamins [4], secondary metabolites [5] and in the activation and detoxification of many xenobiotics such as drugs, carcinogens and environmental pollutants [4,6]. The P450s act as a terminal electron acceptor in the multicomponent P450 dependent monooxygenation system (P450 systems) which leads to the reductive activation of molecular oxygen followed by the insertion of one oxygen atom into the substrate molecule, which catalyzes the following general reaction: RH + O2 + 2e− + 2H+ → ROH + H2O, where R represents the substrate molecule [7]; the required electrons are generally supplied by NADPH and transferred to P450 by a P450 redox partner [8]. In the class II eukaryotic microsomal P450 systems, the general P450 redox partner is a cytochrome P450 reductase, termed CPR [1,7,9], and the electron flow goes from NADPH to FAD to FMN and finally to the heme group in P450. However, alternative electron transfer mechanisms from NADH via cytochrome b5 reductase and cytochrome b5, which are anchored to the endoplasmic reticulum (CBR and CYB5), have been reported [10]. In this last case, the electron flow goes from NADH to CBR to CYB5 and finally to P450, stimulating and increasing P450 reactions [2,11]. Particularly in the fungus Phanerochaete chrysosporium, CBR and CYB5 are involved in the function of the P450 CYP63A2 that is capable of oxidizing polycyclic aromatic hydrocarbons, alkyl phenols and alkanes [12]. In Saccharomyces cerevisiae it was also demonstrated that CBR and CYB5 could maintain CYP51 activity [13]. In cpr - mutant strains of the ascomycete Fusarium fujikuroi, the P450s activities involved in gibberellin biosynthesis showed changes in their regioselectivity, reaction rate and formation of alternative products due to their interaction with alternative redox partners [14].
The basidiomycete yeast Xanthophyllomyces dendrorhous has an enzyme related to the 3A sub-family member cytochrome P450, astaxanthin synthase CrtS (encoded by the crtS gene), which converts beta-carotene into astaxanthin [15,16]. This carotenoid has huge biotechnological potential, as it is currently use in the pharmaceutical and cosmetic industries due to its antioxidant properties and in aquiculture for salmonid fish pigmentation [17]. Astaxanthin synthase has not been reported in other astaxanthin-producing organisms, suggesting that in X. dendrorhous yeast, a unique P450 system has evolved that is specialized in the synthesis of astaxanthin [16].
In addition to astaxanthin synthase, other two P450 encoding genes have been described in X. dendrorhous that are involved in ergosterol biosynthesis: CYP61 [18] and CYP51 [19]. Sterols are essential structural and regulatory components of eukaryotic cell membranes that modulate their thickness, fluidity and permeability [20]; ergosterol is the principal sterol in yeasts that fulfills similar functions as cholesterol in mammalian cells. Although in most organisms there are several P450 encoding genes, in most species, there is only one CPR encoding gene, with few exceptions [21]. The X. dendrorhous CPR gene (named as crtR in this yeast) was characterized and was shown to be essential for the synthesis of astaxanthin [22]. It has been proposed that the X. dendrorhous astaxanthin producing cytochrome P450 system (CrtS and CrtR) is also unique, because CrtS has a high specificity for its own CPR, CrtR. The astaxanthin production in metabolically engineered Saccharomyces cerevisiae strains was only achieved when crtS was co-expressed with crtR [23]. Furthermore, according to protein modeling and molecular dynamics simulations, it was predicted that CrtS interacts preferentially with the FMN-binding domain of CrtR rather than with the one of the CPR of S. cerevisiae, due to a larger interfacial area of interaction and a higher number of hydrogen bonds and saline bridges formed at the interaction surface [24].
Considering the relevance of sterols for the cell, the fact that crtR gene disruption is not lethal in X. dendrorhous [22], indicates the existence of alternative cytochrome P450 electron donors. The aim of this study was to identify and characterize an alternative to the CPR P450 electron donor, specifically the CBR-CYB5 pathway in X. dendrorhous. Considering that unlike CPR, CBR is preferentially NADH-dependent, cbr - mutants were constructed and analyzed. The obtained results provide evidence that the CBR-CYB5 pathway partially complements the crtR - mutation by fulfilling an alternative P450 redox-partner role.
Materials and Methods
Strains and culture conditions
All strains used and constructed in this work are listed in Table 1. The X. dendrorhous crtR - mutant strain, CBSTr [22] that derives from the wild-type strain CBS 6938 (ATCC 96594), were used in this work for phenotypic analyses and genetic modifications. The original X. dendrorhous genomic and transcriptomic sequences were obtained from strain UCD 67–385 (ATCC 24230) by two Next Generation Sequencing (NGS) platforms [25]. Additionally, the genome from strain CBS 6938 was released this year [26].
Table 1. Strains and plasmids used and/or constructed in this work.
| Genotype or Relevant Feature | Reference or Source | |
|---|---|---|
| Strains: | ||
| E. coli | ||
| DH-5alpha | F- φ80d lacZΔM15Δ (lacZY-argF) U169 deoR recA1 endA1 hsdR17(rk- mk+) phoA supE44l- thi-1 gyrA96 relA1. | [35] |
| X. dendrorhous | ||
| UCD 67–385 | ATCC 24230, wild-type. Diploid strain [55]. | ATCC |
| CBS 6938 | ATCC 96594, wild type. | ATCC |
| CBSTr | crtR - mutant strain from CBS 6938 parental wild-type strain. Hygromycin B resistant mutant. | [22] |
| CBS-cbr.1- | cbr.1- mutant strain from CBS 6938 parental wild-type strain. Hygromycin B resistant mutant. | This work |
| CBS-cbr.2- | cbr.2- mutant strain from CBS 6938 parental wild-type strain. Zeocin resistant mutant. | This work |
| Plasmids: | ||
| pBluescript SK- (pBS) | ColE1 ori; AmpR; cloning vector with blue-white selection. | Stratagene |
| pMN-hph | pBS contained at the EcoRV site a cassette of 1.8 kb bearing the E. coli-Hygromycin B resistance (hph) gene under EF-1 α promoter and the GPD transcription terminator of X. dendrorhous. | [56] |
| pIR-zeo | pBS contained at the EcoRV site a cassette of 1.2 kb bearing the Streptoalloteichus hindustanus Zeocin resistance Sh ble gene under the EF-1 α promoter and GPD transcription terminator of X. dendrorhous. | [18] |
| pXd-gCBR.1::hph | pBS contained at the EcoRV, 865 pb upstream, 750 pb downstream of the CBR.1 gene and the hygromycin B resistance cassette between them. | This work |
| pXd-gCBR.2::zeo | pBS contained at the EcoRV, 677 pb upstream, 803 pb downstream of the CBR.2 gene and the zeocin resistance cassette between them. | This work |
ATCC: American Type Culture Collection.
For most experiments, strains CBS 6938, CBSTr, CBS-cbr.1::hph and CBS-cbr.2::zeo were cultured independently in three replicates, which were incubated at 22°C with constant agitation in 1 L Erlenmeyer flasks containing 400 mL of YM medium (1% glucose, 0.3% yeast extract, 0.3% malt extract and 0.5% peptone). The growth curves of each strain were constructed by registering the optical density of the cultures at 600 nm with a V-630 UV-Vis Spectrophotometer from JASCO. For the analyses, 65 mL samples were taken for: i) yeast dry weigh determination (2 samples of 5 mL each), ii) carotenoid extraction (one sample of 15 mL), iii) sterol extraction (one sample of 5 mL), iv) obtaining the microsomal fraction (2 samples of 10 mL) for cytochrome c reductase activity assays and v) RNA extraction (4 samples of 5 mL each). The obtained cellular pellets were washed with sterile distilled water and kept at -80°C until the samples were processed.
Carotenoid and Sterol extraction, and RP-HPLC analyses
Carotenoids and sterols were extracted according to [27] and [28], respectively, quantified spectrophotometrically and normalized to the dry weight of the yeast. Carotenoids were quantified at 465 nm using an absorption coefficient of A1% = 2,100 and sterols at 280 nm were quantified using an absorption coefficient of A1% = 11,500. The extracted carotenoids and sterols were separated by RP-HPLC using an RP-18 Lichrocart 125–4 (Merck) column with acetonitrile: methanol: isopropanol (85:10:5, v/v) and methanol: water (97:3, v/v) as the mobile phase, with a 1 mL/min flux under isocratic conditions. The elution spectra were recovered using a diode array detector; carotenoids and sterols were identified according to their spectra and retention time compared to standards.
Obtaining the microsomal fraction
The microsomal fraction was obtained from cell pellets from 10 mL of culture suspended in 4 mL of extraction buffer (1 mM EDTA, 50 mM Tris HCl pH 7.5, 1 mM DTT, 0.3 M Sorbitol, CompleteTM protease inhibitor cocktail from Boehringer Mannheim). Cells were lysed mechanically by sonication using a Cole Parmer 4710 ultrasonic homogenizer with 500 μL of 0.5 mm glass beads (BioSpec). Twenty sonication pulses of 20 s at 4°C, followed by a 1 min incubation on ice. The cell lysate was centrifuged at 1,000 x g for 3 min and the supernatant was recovered and centrifuged at 20,000 x g at 4°C for 30 min; the new supernatant was recovered and centrifuged at 100,000 x g at 4°C for 1 h. The pellet obtained was suspended in wash buffer (1 mM EDTA, 50 mM Tris HCl pH 7.5, 1 mM DTT, and 0.3 M sorbitol) and centrifuged at 100,000 x g for 1 h at 4°C. Finally, the obtained pellet, which corresponded to the microsomal fraction, was suspended in 200 μL of buffer (30 mM potassium phosphate pH 7.8 and 0.1 mM EDTA) and fractioned into 40 μL samples in Eppendorf tubes, which were frozen with liquid nitrogen and stored at -80°C until use. The protein concentration was determined using the Coomassie Plus Assay Kit (Thermo Scientific) according to the supplier.
Cytochrome c reductase activity assay
The methodology used was adapted from [29], which measured the cytochrome c absorbance increment at 550 nm when it was reduced. The assay was performed in a 275 μL cuvette (UV-Micro, BRAND) containing 237.5 μL assay solution (260 mM potassium phosphate pH 7.8, 0.09 mM EDTA, BSA 0.01 mg/mL and 31 mM cytochrome c) and 5 to 10 μL of the microsomal fraction sample. The reaction was started by adding 12.5 μL of NADPH (2 mM) or NADH (2 mM) and the absorbance at 550 nm was registered every 1 s during 5 min using the kinetic mode of the V-630 UV-Vis Spectrophotometer from JASCO at 22 ± 1°C. The initial velocities of each progress curve were determined with the JASCO Spectra Analysis software.
Identification and characterization of the putative CBR encoding gene from X. dendrorhous
The CBR genomic and cDNA sequences from X. dendrorhous were identified by BLAST analyses over the collection of genomic and transcriptomic contigs and scaffolds of strain UCD 67–385 using the homologous genes from Saccharomyces cerevisiae, Phanerochaete chrysosporium and Cryptococcus neoformans as queries [GenBank: Z28365.1, AY835609.1 and XM_569177.1, respectively]. The nucleotide and deduced amino acid sequences were analyzed with the CLC Genomic Workbench, Geneious 6.0.4, and programs available on-line such as: InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/ [30]), Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html [31]) and JPRED 3 (http://www.compbio.dundee.ac.uk/www-jpred/ [32]). Phylogenetic analyses were carried out using MEGA v.6.06 [33] with the maximum-parsimony method and 1,000 bootstrap replicates.
DNA extraction, amplification and sequencing
Total DNA from X. dendrorhous was extracted according to [34]. All oligonucleotides used in this study were listed in S1 Table and were purchased at Integrated DNA Technologies (IDT). PCR reactions were performed in a 2720 (Applied Biosystems) thermal cycler at a final volume of 25 μL containing 2 U of Pfu DNA polymerase (Thermo Scientific), 2.5 μL of 10X Pfu buffer, 0.5 μL of 10 mM dNTPs, 1 μL of 50 mM MgCl2, 1 μl of each primer (25 μM) and 10 to 20 ng of template DNA. In general, the amplification protocol was: initial denaturation at 95°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and synthesis at 72°C for 3 min; and a final extension step at 72°C for 10 min. Samples were kept at 4°C until analysis by 0.8% agarose gel electrophoresis in TAE buffer containing 0.5 μg/mL ethidium bromide [35]. DNA for sequencing was purified from gels by the glass milk method [36]. Nucleotide sequences were obtained from an ABI 3100 Avant genetic analyzer using the BigDye terminator v3.1 kit (Applied Biosystems).
RNA extraction, single strand DNA synthesis and quantitative PCR (RT-qPCR)
Total RNA was extracted according to a modified protocol from Chomczynski and Sacchi [37,38] and RNA was quantified spectrophotometrically at 260 nm [35]. The synthesis of cDNA was performed according to the M-MLV reverse transcriptase (Invitrogen) manufacturer’s protocol, with 5 μg of total RNA at a final volume of 20 μL using oligo-dT18. The relative transcript level determination was performed in an Mx3000P quantitative PCR system (Stratagene) using primers pairs (S1 Table) with efficiencies greater than 95%, as determined by standard curves with a correlation coefficient of R2 ≥ 0.996. Each reaction contained 1 μL of the reverse transcription reaction, 0.25 μM of each primer and 10 μL of the SensiMix SYBR Green I (Quantace) kit at a final volume of 20 μL. The obtained Ct values were normalized to the respective value of beta-actin [GenBank: X89898.1] [39] and were later expressed as a function of the control conditions (wild type strain) using the ΔΔCt algorithm [40].
Plasmid construction and X. dendrorhous transformation
All plasmids used in this work were listed in Table 1. To knockout the X. dendrorhous CBR genes, the plasmids pXd-gCBR.1::hph and pXd-gCBR.2::zeo were constructed. Initially, the upstream and downstream DNA region of approximately 850 pb for each gene were PCR-amplified from genomic DNA from strain UCD 67–385 using the primers pairs pre_gCBR1-Fw + gCBR1-HpaI-Rv, gCBR1-HpaI-Fw + post_gCBR1-Rv, cytb5Red_del-Fw + cytb5Red_del_HpaI-Rv, and cytb5Red_del_HpaI-Fw + cytb5Red_del-Rv to amplify the CBR.1 upstream, the CBR.1 downstream, the CBR.2 upstream and the CBR.2 downstream regions, respectively. Then, the upstream and downstream DNA regions of each gene were joined by OE-PCR [41] leaving an HpaI restriction site between them according to the design of the primers that were used. The new DNA fragments were ligated into the EcoRV site of the plasmid pBluescript SK- [35]. Finally, the plasmids pXd-gCBR.1::hph and pXd-gCBR.2::zeo were generated by digesting the previous plasmids with HpaI and introducing an antibiotic resistance cassette (hygromycin B: hph or zeocin: zeo) for X. dendrorhous transformant selection.
Electrocompetent X. dendrorhous cells obtained from exponential cultures (OD600nm = 1.2) grown in YM medium were transformed by electroporation using a Bio-Rad gene pulser X cell with PC and CE modules under the following conditions: 125 mF, 600 Ω, and 0.45 kV. Transformations were performed using 1 to 5 μg of linear donor DNA, which was released from the plasmids pXd-gCBR.1::hph and pXd-gCBR.2::zeo by digesting with NotI + ClaI and NotI + XhoI, respectively. Yeast transformants were selected on YM-agar plates (1.5%) supplemented with 35 μg/mL hygromycin-B or 40 μg/mL zeocin when necessary.
Results and Discussion
Phenotypic analysis of the wild-type and crtR - mutant X. dendrorhous strains
In a previous work, the gene that encodes the X. dendrorhous CPR (CrtR, crtR gene) was identified and was demonstrated to be essential in the production of astaxanthin in this yeast [22]. However, its participation in other pathways involving P450 enzymes, such as ergosterol biosynthesis, was not analyzed. As such, we performed phenotypic analyses focusing on the production of astaxanthin (as control) and ergosterol in the wild-type (CBS 6938) and crtR - mutant strain (CBSTr) derived from CBS 6938. Both strains were cultivated in triplicate and samples were taken along the growth curve (after 21, 38, 72 and 120 h of culture, corresponding to early exponential, middle exponential, early stationary and late stationary phases of growth, respectively) to evaluate the production of carotenoids and sterols. Statistical analyses (Student’s t test, p<0.05) confirmed that the CBSTr strain reached a higher carotenoid content than the wild-type strain after 120 h of culture. However, as was previously reported, CBSTr does not produce astaxanthin but accumulates beta-carotene, indicating that only CrtR supports the electron flow to astaxanthin synthase (CrtS), which belongs to the P450 protein family.
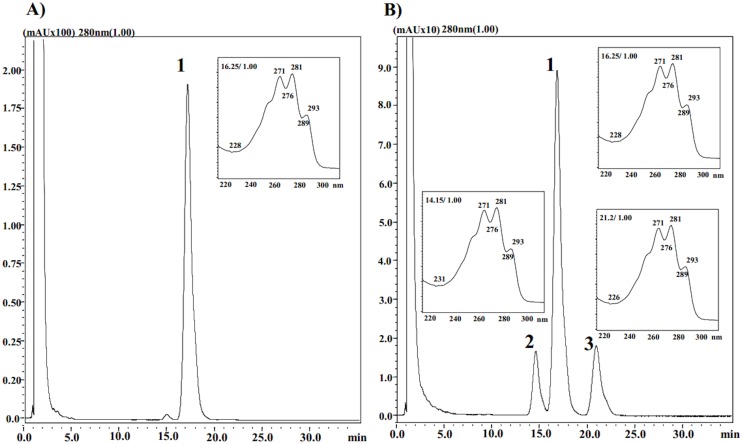
In contrast, total sterol production was similar between both strains at all analyzed culture times, but the sterol composition was different. All the sterol samples from the wild-type strain showed a single predominant RP-HPLC peak at the 280 nm channel, with a retention time of approximately 16 min (peak 1) with the ergosterol characteristic UV-spectra (Fig 1A). The identity of this sterol was confirmed by co-injecting each sample with standard ergosterol. On the other hand, three peaks with sterol characteristic UV-spectra at retention times of approximately 15 (peak 2), 16 (peak 1) and 22 (peak 3) min were observed in the RP-HPLC profile of sterol samples from the crtR - mutant strain, with ergosterol (peak 1, also confirmed by co-injecting the samples with standard ergosterol) being the most abundant (Fig 1B). These results show that in X. dendrorhous, there must be an alternative to the CPR P450 electron donor to sustain the synthesis of ergosterol in CBSTr, although other sterols beside ergosterol are accumulated. This was not the case in S. cerevisiae, as the total sterol content in a cytochrome P450 reductase mutant strain was approximately 4-fold lower than in the wild-type strain, but the ergosterol fraction in both strains was the same (approximately 90%) [42]. Interestingly, the retention times of peaks 2 and 3 coincided with the two single peaks observed in the sterol samples from the X. dendrorhous cyp61 - mutant strains [18]. To confirm this observation, sterol samples were obtained from the crtR - and cyp61 - mutant strains which were independently co-injected with standard ergosterol and also mixed together, revealing that CBSTr indeed accumulates the same sterols as the cyp61 - mutant, besides ergosterol. This result suggests that the functionality of the CYP61 enzyme could be partially affected in the CBSTr mutant strain. However, this strain still produces ergosterol, so there must be a P450 electron donor via an alternative to cytochrome P450 reductase in X. dendrorhous that complements ergosterol, but not astaxanthin biosynthesis. The results indicated that the alternative electron donor, probably CBR-CYB5, had a different affinity for the P450s enzymes CrtS, CYP51 and CYP61. This agrees with in vitro enzymatic assays described in S. cerevisiae CBR, CYB5 and CYP51, in which it was determined that CYP51 had efficient activity with CBR-CYB5 [13].
Fig 1. RP-HPLC analysis of sterols from the wild-type and crtR- mutant strain.
Chromatograms (at 280 nm) correspond to sterols extracted after 72 h of culture from the (A) wild-type (CBS 6938) and (B) CBSTr strain (crtR -). The corresponding spectra were displayed beside each peak (peaks N° 1 to 3). Peak 1 corresponds to ergosterol, which was confirmed by co-injecting the sample with standard ergosterol.
Identification and molecular characterization of the X. dendrorhous CBR and CYB5 genes
By BLAST analyses over the genomic and transcriptomic sequences of the UCD 67–385 strain, we were able to identify the putative X. dendrorhous CBR and CYB5 genes. Two CBR and a single CYB5 genes were identified, which were named CBR.1, CBR.2 and CYB5, respectively, and were uploaded to the GenBank database [KT448554, KT448556 and KT448555, respectively]. From the sequence analyses of the gDNA and cDNA versions of each gene, their genetic structures were determined.
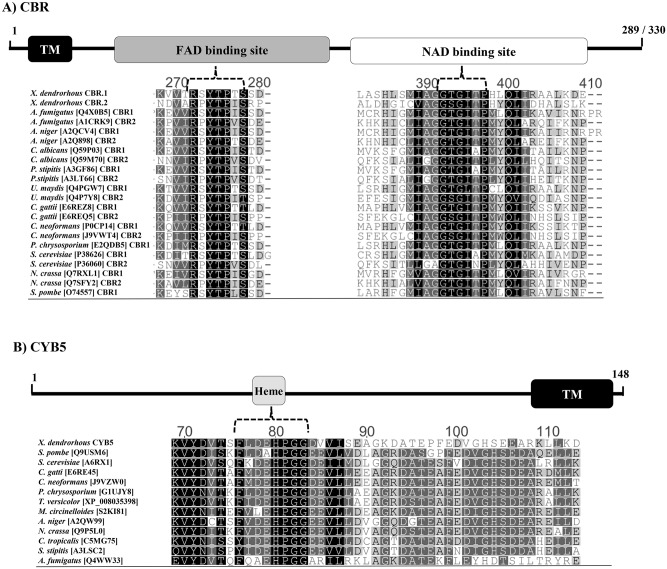
The X. dendrorhous CBR.1 gene has an ORF of 870 bp and consists of 7 exons of 103, 85, 316, 82, 35, 173, 76, bp and 6 introns of 123, 68, 89, 93, 97 and 132 bp. This gene encodes a predicted CBR protein of 289 amino acids with a molecular weight of 31.48 kDa (pI, 6.96). In contrast, the CBR.2 gene has 9 exons of 82, 12, 121, 307, 111, 154, 92, 40 and 74 bp, and 8 introns of 247, 126, 104, 92, 95, 86, 121 and 87 bp, giving an ORF of 993 bp. The X. dendrorhous CBR.2 gene encodes a predicted 330 amino acid protein with molecular weight of 35.35 kDa (pI, 8.41). Bioinformatic analyses of both deduced X. dendrorhous CBR proteins also revealed structurally conserved elements in the cytochrome b5 reductase protein family: i) an NADH binding domain (signature: G-x-G-x-x-P), ii) an FAD binding domain (signature: R-x-Y-T-x-x-S) and iii) a transmembrane segment at the amino terminal region, through which CBRs anchors to the endoplasmic reticulum (Fig 2A). Interestingly, the subcellular distribution of both X. dendrorhous CBR proteins might be different according to the PSORT tool based on the method of McGeoch's [43], which predicted that CBR.1 was likely located in the endoplasmic reticulum, while CBR-2 was located in the mitochondrial membrane. Moreover, by a phylogenetic analysis, it was found that the CBR.1 groups together with other CBRs were most of them have been described as microsomal, while CBR.2 showed a closer relation with CBRs described as mitochondrial (Fig 3). For these reasons, it was most likely that CBR.1, and not CBR.2, would be involved in the class II P450 systems in X. dendrorhous.
Fig 2. X. dendrorhous CBR.1, CBR.2 and CYB5 protein amino acid sequence alignment.
Amino acid sequence alignment of conserved protein motives: (A) CBR and (B) CYB5. The X. dendrorhous CBR.1, CBR.2 and CYB5 are included. The UniProtKB or GenBank accession number is indicated in brackets beside the organism name and above each alignment; a scheme of the deduced X. dendrorhous protein structure is included. Tm: trans-membrane segment.
Fig 3. Phylogenetic analysis of the CBR genes from X. dendrorhous.
The accession numbers for each of the amino acid sequences are: Cryptococcus gattii CBR1 [E6REZ8] and CBR2 [E6REQ5], C. neoformans CBR1 [P0CP14] and CBR2 [J9VWT4], P. chrysosporium CBR1 [E2QDB5], T. versicolor CBR1 [XP_008033072], U. maydis CBR1 [Q4PGW7] and CBR2 [Q4P7Y8], P. stipitis CBR1 [A3GF86] and CBR2 [A3LT66], C. albicans CBR1 [Q59P03] and CBR2 [Q59M70], N. crassa CBR1 [Q7RXL1] and CBR2 [Q7SFY2], A. niger CBR1 [A2QCV4] and CBR2 [A2Q898], A. fumigatus CBR1 [Q4X0B5] and CBR2 [A1CRK9], S. pombe CBR1 [O74557], S. cerevisiae CBR1 [P38626] and CBR2 [P36060], V. vinifera CBR1 [F6HIY1], T. urartu CBR1 [M7YU08], A. thaliana CBR1 [Q9ZNT1], H. armigera CBR1 [F1CZW3], D. rerio CBR1 [Q7ZVF8], S. scrofa CBR1 [F1S4N2], H. sapiens CBR1 [Q9UHQ9], M. musculus CBR1 [Q9DB73], and R. norvegicus CBR1 [G3V9S0]. The UniProtKB or GenBank accession number is indicated in brackets. The value at each node indicates the Bootstrap after 1000 iterations using MEGA v.6.06.
Similarly, the CYB5 gene has an ORF of 444 bp with 6 exons of 145, 39, 27, 95, 68 and 73 bp, and 5 introns of 227, 105, 115, 87 and 110 bp. This gene encodes a 148 amino acid polypeptide with a predicted molecular weight of 16.07 kDa (pI, 4.87). The deduced protein size was consistent with CYB5 proteins described in other fungi, given that the reported size of this protein ranged from 120 to 158 amino acids [44–46]. Moreover, structural conserved elements in the cytochrome b5 protein family were identified in the deduced amino acid structure of the X. dendrorhous CYB5 gene including: i) a putative hydrophobic transmembrane segment at the carboxyl terminus, which could anchor the protein to the endoplasmic reticulum and ii) the cytochrome b5 family heme-binding domain with the conserved signature: [FY]-[LIVMK]-(I)-(Q)-H-P-[GA]-G [47] (Fig 2B).
CBR.1 and CBR.2 gene mutations in X. dendrorhous
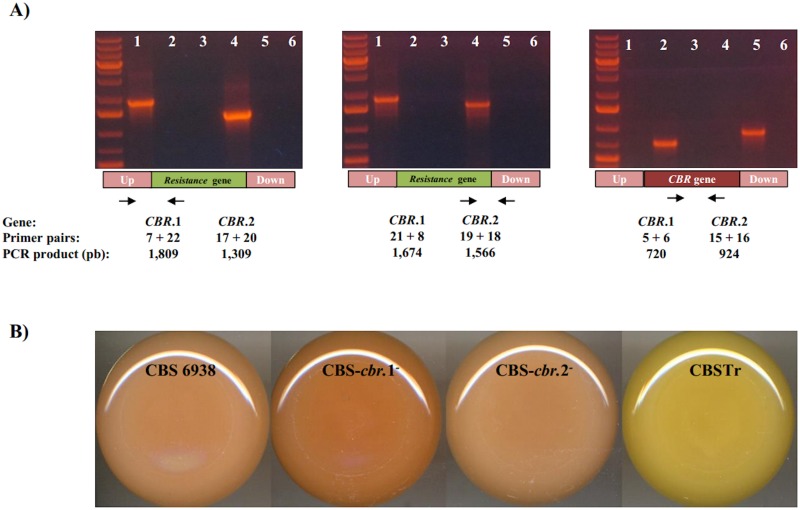
As CBR is the enzyme responsible for the capture of electrons directly from NADH, it was expected that CBR.1 or CBR.2 gene mutations would decrease the CBR-CYB5 electron supply by using NADH as a cofactor. The mutant strains CBS-cbr.1::hph and CBS-cbr.2::zeo were obtained from the wild-type strain CBS 6938, where the corresponding gene was replaced with an antibiotic resistance marker through a double homologous recombination event (Table 1). According to previous results [22], the wild-type CBS 6938 strain was likely aneuploid, and the CBS-cbr.1::hph and CBS-cbr.2::zeo strains were hemizygous as PCR-based genotype analysis revealed that a unique CBR gene copy was mutated in each case (Fig 4). We evaluated the effect of both mutations on carotenoid and sterol production at two points of the growth curve, and for comparative purposes, the wild-type and the crtR - mutant strains were included, which were cultured in parallel with the other two strains; the results were summarized in Table 2. The CBS-cbr.1- strain carotenoid production was 3-fold higher than the wild-type strain at 72 h, while strain CBS-cbr.2- did not show significant differences; in both mutant strains, the carotenoid composition was similar to that of the parental strain, and astaxanthin was the main carotenoid. The two cbr- mutants and the wild-type strain showed a similar sterol production and composition at both culture times, with ergosterol (approximately 98%) being the main sterol. These results indicate that even though one of the two CBR genes products could be involved in ergosterol biosynthesis, crtR complemented the absence of the CBR gene in the mutant strains, at least for ergosterol production. It would be very informative to obtain a crtR and CBR.1 double mutant strain; however, this might not be possible as in S. cerevisiae it has been shown that the double mutant of the cytochrome P450 reductase and cytochrome b5 encoding genes is not viable [48]. Considering this and to confirm if CrtR and CBR.1 mediate the oxidation of sterols, future in vitro enzymatic reaction experiments with CYP51 or CYP61 coupled with CrtR and CBR.1 could be performed.
Fig 4. PCR-based analysis of the CBS-cbr.1- and CBS-cbr.2- strains.
(A) Each gel shows the PCR reaction products obtained with different primers pairs numbered according to S1 Table and, as template, genomic DNA from the following strains: CBS-cbr.1- (line 1), CBS 6938 (line 2), negative primer control (line 3), CBS-cbr.2- (line 4), CBS 6938 (line 5), and negative primer control (line 6). Under each gel, a general scheme of the PCR analyses is included representing the resistance cassettes (green), the corresponding CBR gene (red) and the CBR flanking DNA (pink). A 1 kb plus DNA ladder (20.0, 10.0, 7.0, 5.0, 4.0, 3.0, 2.0, 1.5, 1.0, 0.7, and 0.5 Kbp) was used as the molecular weight standard in the right lane of each gel. (B) Color phenotype of the strains CBS 6938 (WT), CBS-cbr.1-, CBS-cbr.2- and CBSTr after 5 days of culture on YM medium incubated at 22°C with constant agitation.
Table 2. Carotenoids and sterols produced by the X. dendrorhous strains cultured in parallel after 36 and 72 h of cultivation.
| Metabolite production | ||||
|---|---|---|---|---|
| Strain | Total carotenoids (μg carotenoids/g dry yeast weight) |
Total sterols (mg sterols/g dry yeast weight) |
||
| 36 h | 72 h | 36 h | 72 h | |
| CBS 6938 | 57.5 ± 4.7 | 62.4 ± 3.5 | 7.8 ± 0.4 | 4.0 ± 0.6 |
| CBSTr | 46.4 ± 8.3 | 82.9 ± 4.4* | 6.0 ± 1.0 | 4.0 ± 0.3 |
| CBS-cbr.1- | 61.0 ± 16.4 | 224.2 ± 4.5* | 5.8 ± 0.3 | 4.2 ± 0.3 |
| CBS-cbr.2- | 54.2 ± 18.6 | 91.0 ± 21.3 | 5.5 ± 0.1 | 3.9 ± 0.4 |
* Statistically significant differences between the wild-type and mutant strains (Student’s t test, p<0.05 and α = 0.05).
Cytochrome c reductase activity and gene expression in mutants and wild-type strains
The cytochrome c reductase activity assay is a useful approximation to evaluate cytochrome P450 reductase activity with NADPH as a reductant, as cytochrome c is an artificial substrate for CPR. Additionally, it has been shown that in some higher eukaryotes, CBR reduced cytochrome c with NADH only in the presence of CYB5 [49,50]; in others cases, it may do this directly [51,52]. Therefore, even though the cytochrome c reductase assay may not encompass the functionality of the complete CBR-CYB5 electron donor pathway, it is a useful approximation to determine cofactor preference in our mutants. In this way, indications of the contribution of each electron transfer via: NADPH or NADH for CPR or CBR-CYB5, respectively, can be obtained to determine if they are operative in X. dendrorhous microsomal extracts.
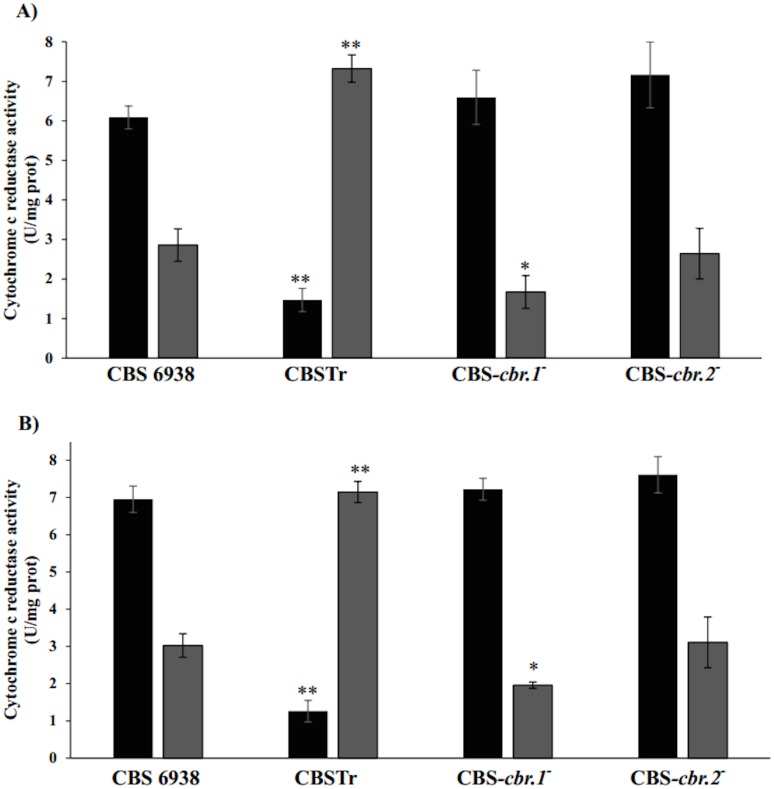
To evaluate the functionality of these enzymes in X. dendrorhous, cytochrome c reductase activity assays were performed with microsomal extracts from crtR - (CBSTr), CBS-cbr.1-, and CBS-cbr.2- mutants and the wild-type strain using NADPH or NADH as cofactors. The microsomal fraction was obtained after 36 and 72 h of cultivation from the same three replica cultures used for metabolite production analyses in the four strains; the results were summarized in (Fig 5). In general, the microsomal fraction obtained from the wild-type strain at all cultivation times showed a higher cytochrome c reductase activity with NADPH than with NADH as a cofactor. However, the microsomal fractions from strain CBSTr showed an inverse pattern; they had higher cytochrome c reductase activity with NADH than with NADPH. Moreover, by comparing the activity results between these two strains, microsomes from CBSTr showed on average approximately 3-fold less activity with NADPH, whereas with NADH, the activity was almost 2-fold higher in relation to the wild-type strain. These are interesting results revealing that in order to reduce cytochrome c, microsomes from the wild-type strain have a preference for NADPH, while samples from CBSTr have a preference for NADH, which agrees with the absence of CPR. Similar to the wild-type strain, microsomes from the CBS-cbr.1- and CBS-cbr.2- mutant strains also showed higher cytochrome c reductase activity with NADPH than with NADH; however, microsomes from CBS-cbr.1- showed lower NADH-dependent cytochrome c reductase activity in relation to the wild-type strain, while microsomes from CBS-cbr.2- did not show significant differences (Student’s t test, p<0.05). As a whole, these results suggest that crtR in the CBS-cbr.1- and CBS-cbr.2- mutant strains contributes by sustaining the NADPH- dependent cytochrome c reductase activity, which could be sufficient to maintain the wild-type ergosterol production level. However, the reduced NADH-dependent cytochrome c reductase activity in CBS-cbr.1- strongly suggests that CBR.1, and not CBR.2, is involved in the X. dendrorhous class II P450 systems, supporting the results from the bioinformatics analyses.
Fig 5. Cytochrome c reductase activity of the wild-type and mutant strains.
Assays were performed with NADPH (black bars) or NADH (grey bars) as cofactor using microsomal fractions extracted after (A) 36 h and (B) 72 h of culture. Values are the mean ± standard deviation of two technical replicates from three independent cultures. Statistical significant differences between wild-type and mutant strain compared to the same assay are indicated (Student’s t test, *P<0.05, **P<0.01 with α = 0.05).
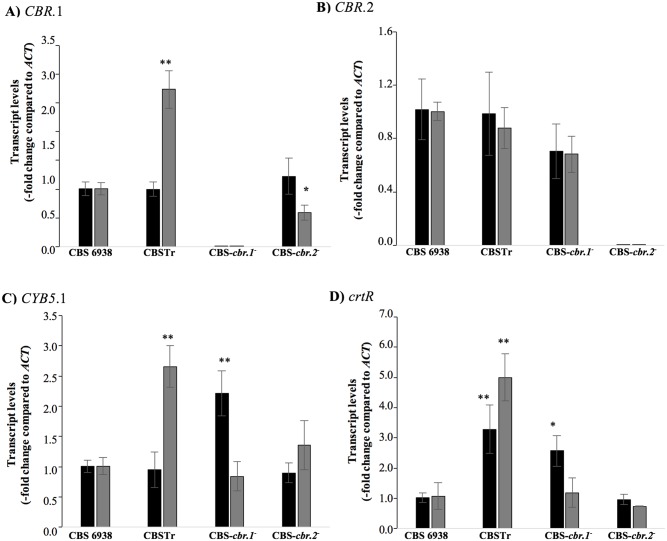
Considering the higher NADH- and the lower NADPH-dependent cytochrome c reductase activity in the crtR - mutant strain in relation to the wild-type strain, we evaluated if this could be a consequence of differential transcript levels of the studied genes among the analyzed strains in this work. Total RNA was extracted at the same time-points of the same cultures used in the previous studies, and crtR, CBR.1, CBR.2 and CYB5 transcript levels were evaluated by RT-qPCR (Fig 6). As expected, the CBS-cbr.1- and CBS-cbr.2- strains did not show CBR.1 and CBR.2 transcripts. The transcript levels of the CBR.2 gene did not show significant differences between the analyzed strains, which was consistent with the previous results obtained in this work suggesting that the CBR.2 gene is not involved in the class II P450 systems. In the CBS-cbr.2- strain, only CBR.1 showed a lower transcript level in relation to the wild-type strain after 72 h of culture. Interestingly, even though no differences were detected at the early stationary phase (36 h) of growth, the higher NADH-dependent cytochrome c reductase activity observed in microsomes obtained from CBSTr correlated with higher CBR.1 and CYB5 transcript levels in relation to the wild-type strain after 72 h of culture. This observation could be explained as a compensatory mechanism for cytochrome P450 activity due to the crtR gene mutation in the CBSTr strain. This idea was also supported by the fact that there was a higher NADH-dependent cytochrome c reductase activity in the CBSTr strain, suggesting that in the absence of a functional CPR, the activity of an alternative P450 electron donor, likely CBR-CYB5 via, was enhanced in X. dendrorhous.
Fig 6. RT-qPCR for CBR.1. CBR.2, CYB5 and crtR transcript levels in the wild-type and mutant strains.
(A) CBR.1 (B) CBR.2, (C) CYB5 and (D) crtR genes. The transcript level of each gene was normalized to actin mRNA. Black and grey bars represent the results using RNA extracted after 36 and 72 h of culture, respectively. Values are the mean ± standard deviation of two technical replicates from three independent cultures (Student’s t-test was performed to compare each mutant to the parental strain. * P <0.05 and ** P <0.01).
Finally, the transcript level of the crtR gene was determined. This gene was not completely deleted in the CBSTr strain, but it was interrupted with a module that confers resistance to hygromycin B [22]. Even though the gene product was not functional, it can be transcribed, allowing us to include it in these analyses. In the two culture time-points of growth analyzed, the crtR transcript levels in CBSTr were higher than in the wild-type strain. Considering that the sterol composition was altered in this strain, the reduced ergosterol content could enhance crtR gene expression as has been reported for other genes [18]. Interestingly, the transcript level of this gene was also higher than the wild-type strain in the CBS-cbr.1- strain after 36 h of culture, suggesting that crtR could compensate for the absence of the CBR.1 gene in cytochrome P450 activity. Previously it was reported that the regulation of the expression of the cytochrome P450 reductase gene, which encodes the main electron donor in P450s systems, was particularly complex and involved several regulatory elements in the promoter region, differential promoter use and regulation at the post-transcriptional level [53,54]. As such, it was expected that the regulation of alternative redox partners such as CBR-CYB5 could also be subject to complex regulatory mechanisms. However, the mechanism by which the crtR - gene mutation affects the CYB5 and CBR.1 transcript levels requires further investigation.
Conclusions
In addition to being essential for astaxanthin biosynthesis, our results show that the X. dendrorhous crtR gene is also involved in ergosterol biosynthesis, as the ergosterol proportion was reduced in the crtR - mutant. The higher transcript level of the CBR.1 and CYB5 genes and the increased NADH-dependent cytochrome c reductase activity in the crtR - mutant strain, as well as the reduced NADH-dependent cytochrome c reductase activity in the CBS-cbr.1- mutant strain, all strongly suggest the involvement of CBR.1-CYB5 as an alternative electron donor to P450 enzymes during sterol biosynthesis in X. dendrorhous.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s website.
(PDF)
Acknowledgments
This work was financially supported by FONDECYT 11121200 to JA and by a CONICYT Doctoral Fellowship to MSG.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Fondo Nacional de Desarrollo Cientifico y Tecnologico 11121200, (http://www.conicyt.cl/fondecyt/), to JA, and Comision Nacional de Investigacion Cientifica y Tecnologica 21130807 (http://www.conicyt.cl/), to MSG.
References
- 1. McLean KJ, Sabri M, Marshall KR, Lawson RJ, Lewis DG, Clift D, et al. (2005) Biodiversity of cytochrome P450 redox systems. Biochemical Society Transactions 33: 796–801. [DOI] [PubMed] [Google Scholar]
- 2. Zhang H, Im SC, Waskell L (2007) Cytochrome b5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J Biol Chem 282: 29766–29776. [DOI] [PubMed] [Google Scholar]
- 3. Degtyarenko KN, Archakov AI (1993) Molecular evolution of P450 superfamily and P450-containing monooxygenase systems. FEBS letters 332: 1–8. [DOI] [PubMed] [Google Scholar]
- 4. Bernhardt R (2006) Cytochromes P450 as versatile biocatalysts. J Biotechnol 124: 128–145. [DOI] [PubMed] [Google Scholar]
- 5. Estabrook RW (2003) A passion for P450s (rememberances of the early history of research on cytochrome P450). Drug Metab Dispos 31: 1461–1473. [DOI] [PubMed] [Google Scholar]
- 6. Porter TD, Coon MJ (1991) Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem 266: 13469–13472. [PubMed] [Google Scholar]
- 7. van den Brink HM, van Gorcom RF, van den Hondel CA, Punt PJ (1998) Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol 23: 1–17. [DOI] [PubMed] [Google Scholar]
- 8. Kimmich N, Das A, Sevrioukova I, Meharenna Y, Sligar SG, Poulos TL (2007) Electron transfer between cytochrome P450cin and its FMN-containing redox partner, cindoxin. J Biol Chem 282: 27006–27011. [DOI] [PubMed] [Google Scholar]
- 9. Munro AW, Girvan HM, McLean KJ (2007) Cytochrome P450—redox partner fusion enzymes. Biochim Biophys Acta 1770: 345–359. [DOI] [PubMed] [Google Scholar]
- 10. Subramanian V, Doddapaneni H, Syed K, Yadav JS (2010) P450 redox enzymes in the white rot fungus Phanerochaete chrysosporium: gene transcription, heterologous expression, and activity analysis on the purified proteins. Curr Microbiol 61: 306–314. 10.1007/s00284-010-9612-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Porter TD (2002) The roles of cytochrome b5 in cytochrome P450 reactions. J Biochem Mol Toxicol 16: 311–316. [DOI] [PubMed] [Google Scholar]
- 12. Syed K, Kattamuri C, Thompson TB, Yadav JS (2011) Cytochrome b5 reductase-cytochrome b5 as an active P450 redox enzyme system in Phanerochaete chrysosporium: atypical properties and in vivo evidence of electron transfer capability to CYP63A2. Arch Biochem Biophys 509: 26–32. 10.1016/j.abb.2011.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamb DC, Kelly DE, Manning NJ, Kaderbhai MA, Kelly SL (1999) Biodiversity of the P450 catalytic cycle: yeast cytochrome b 5/NADH cytochrome b 5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett 462: 283–288. [DOI] [PubMed] [Google Scholar]
- 14. Troncoso C, Cárcamo J, Hedden P, Tudzynski B, Rojas MC (2008) Influence of electron transport proteins on the reactions catalyzed by Fusarium fujikuroi gibberellin monooxygenases. Phytochemistry 69: 672–683. [DOI] [PubMed] [Google Scholar]
- 15. Alvarez V, Rodríguez-Sáiz M, de la Fuente JL, Gudiña EJ, Godio RP, Martín JF, et al. (2006) The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of beta-carotene into astaxanthin and other xanthophylls. Fungal Genet Biol 43: 261–272. [DOI] [PubMed] [Google Scholar]
- 16. Ojima K, Breitenbach J, Visser H, Setoguchi Y, Tabata K, Hoshino T, et al. (2006) Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a β-carotene 3-hydroxylase/4-ketolase. Mol Genet Genomics 275: 148–158. [DOI] [PubMed] [Google Scholar]
- 17. Higuera-Ciapara I, Felix-Valenzuela L, Goycoolea FM (2006) Astaxanthin: a review of its chemistry and applications. Crit Rev Food Sci Nutr 46: 185–196. [DOI] [PubMed] [Google Scholar]
- 18. Loto I, Barahona S, Sepúlveda D, Martínez-Moya P, Baeza M, Cifuentes V, et al. (2012) Enhancement of carotenoid production by disrupting the C22-sterol desaturase gene (CYP61) in Xanthophyllomyces dendrorhous . BMC Microbiol 12: 235 10.1186/1471-2180-12-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leiva K, Werner N, Sepúlveda D, Barahona S, Baeza M, Cifuentes V, et al. (2015) Identification and functional characterization of the CYP51 gene from the yeast Xanthophyllomyces dendrorhous that is involved in ergosterol biosynthesis. BMC Microbiol 15: 89 10.1186/s12866-015-0428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Rao R (2010) Beyond ergosterol: Linking pH to antifungal mechanisms. Virulence 1: 551–554. [DOI] [PubMed] [Google Scholar]
- 21. Lah L, Krasevec N, Trontelj P, Komel R (2008) High diversity and complex evolution of fungal cytochrome P450 reductase: cytochrome P450 systems. Fungal Genet Biol 45: 446–458. [DOI] [PubMed] [Google Scholar]
- 22. Alcaíno J, Barahona S, Carmona M, Lozano C, Marcoleta A, Niklitschek M, et al. (2008) Cloning of the cytochrome P450 reductase (crtR) gene and its involvement in the astaxanthin biosynthesis of Xanthophyllomyces dendrorhous . BMC Microbiol 8: 169 10.1186/1471-2180-8-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ukibe K, Hashida K, Yoshida N, Takagi H (2009) Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Appl Environ Microbiol 75: 7205–7211. 10.1128/AEM.01249-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alcaíno J, Fuentealba M, Cabrera R, Baeza M, Cifuentes V (2012) Modeling the interfacial interactions between CrtS and CrtR from Xanthophyllomyces dendrorhous, a P450 system involved in astaxanthin production. J Agric Food Chem 60: 8640–8647. [DOI] [PubMed] [Google Scholar]
- 25. Baeza M, Alcaíno J, Barahona S, Sepúlveda D, Cifuentes V (2015) Codon usage and codon context bias in Xanthophyllomyces dendrorhous . BMC Genomics 16: 293 10.1186/s12864-015-1493-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma R, Gassel S, Steiger S, Xia X, Bauer R, Sandmann G, et al. (2015) The genome of the basal agaricomycete Xanthophyllomyces dendrorhous provides insights into the organization of its acetyl-CoA derived pathways and the evolution of Agaricomycotina. BMC Genomics 16: 233 10.1186/s12864-015-1380-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. An GH, Schuman DB, Johnson EA (1989) Isolation of Phaffia rhodozyma mutants with increased astaxanthin content. Appl Environ Microbiol 55: 116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shang F, Wen S, Wang X, Tan T (2006) Effect of nitrogen limitation on the ergosterol production by fed-batch culture of Saccharomyces cerevisiae . J Biotechnol 122: 285–292. [DOI] [PubMed] [Google Scholar]
- 29. Vermilion JL, Coon MJ (1974) Highly purified detergent-solubilized NADPH-cytochrome P-450 reductase from phenobarbital-induced rat liver microsomes. Biochem Biophys Res Commun 60: 1315–1322. [DOI] [PubMed] [Google Scholar]
- 30. Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hofmann K, Stoffel W (1993) TMbase—A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374: 166. [Google Scholar]
- 32. Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36: W197–201. 10.1093/nar/gkn238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cifuentes V, Hermosilla G, Martinez C, Leon R, Pincheira G, Jiménez A (1997) Genetics and electrophoretic karyotyping of wild-type and astaxanthin mutant strains of Phaffia rhodozyma . Antonie Van Leeuwenhoek 72: 111–117. [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- 36. Boyle JS, Lew AM (1995) An inexpensive alternative to glassmilk for DNA purification. Trends Genet 11: 8 [DOI] [PubMed] [Google Scholar]
- 37. Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- 38. Contreras G, Barahona S, Rojas MC, Baeza M, Cifuentes V, Alcaíno J (2013) Increase in the astaxanthin synthase gene (crtS) dose by in vivo DNA fragment assembly in Xanthophyllomyces dendrorhous . BMC Biotechnol 13: 84 10.1186/1472-6750-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lodato P, Alcaíno J, Barahona S, Niklitschek M, Carmona M, Wozniak A, et al. (2007) Expression of the carotenoid biosynthesis genes in Xanthophyllomyces dendrorhous . Biol Res 40: 73–84. [DOI] [PubMed] [Google Scholar]
- 40. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 41. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- 42. Venkateswarlu K, Kelly DE, Manning NJ, Kelly SL (1998) NADPH cytochrome P-450 oxidoreductase and susceptibility to ketoconazole. Antimicrob Agents Ch 42: 1756–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horton P, Nakai K (1996) A probabilistic classification system for predicting the cellular localization sites of proteins. Proc Int Conf Intell Syst Mol Biol 4: 109–115. [PubMed] [Google Scholar]
- 44. Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus . Nature 438: 1151–1156. [DOI] [PubMed] [Google Scholar]
- 45. Malmstrom L, Riffle M, Strauss CE, Chivian D, Davis TN, Bonneau R, et al. (2007) Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biol 5: e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ichinose H, Wariishi H (2012) Heterologous expression and mechanistic investigation of a fungal cytochrome P450 (CYP5150A2): involvement of alternative redox partners. Arch Biochem Biophys 518: 8–15. 10.1016/j.abb.2011.12.010 [DOI] [PubMed] [Google Scholar]
- 47. de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Truan G, Epinat J-C, Rougeulle C, Cullin C, Pompon D (1994) Cloning and characterization of a yeast cytochrome b 5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene 142: 123–127. [DOI] [PubMed] [Google Scholar]
- 49. Strittmatter P, Velick SF (1957) The purification and properties of microsomal cytochrome reductase. J Biol Chem 228: 785–799. [PubMed] [Google Scholar]
- 50. Arinç E, Çakir D (1999) Simultaneous purification and characterization of cytochrome b5 reductase and cytochrome b5 from sheep liver. Int J Biochem Cell Biol 31: 345–362. [DOI] [PubMed] [Google Scholar]
- 51. Kitajima S, Yasukochi Y, Minakami S (1981) Purification and properties of human erythrocyte membrane NADH-cytochrome b5 reductase. Arch Biochem Biophys 210: 330–339. [DOI] [PubMed] [Google Scholar]
- 52. Bagnaresi P, Basso B, Pupillo P (1997) The NADH-dependent Fe3+-chelate reductases of tomato roots. Planta 202: 427–434. [DOI] [PubMed] [Google Scholar]
- 53. van den Brink JM, Punt PJ, van Gorcom RFM, van Den Hondel CAMJJ (2000) Regulation of expression of the Aspergillus niger benzoate para-hydroxylase cytochrome P450 system. Mol Gen Genet 263: 601–609. [DOI] [PubMed] [Google Scholar]
- 54. Alcaíno J, Romero I, Niklitschek M, Sepúlveda D, Rojas MC, Baeza M, et al. (2014) Functional characterization of the Xanthophyllomyces dendrorhous farnesyl pyrophosphate synthase and geranylgeranyl pyrophosphate synthase encoding genes that are involved in the synthesis of isoprenoid precursors. PLoS One 9: e96626 10.1371/journal.pone.0096626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hermosilla G, Martinez C, Retamales P, Leon R, Cifuentes V (2003) Genetic determination of ploidy level in Xanthophyllomyces dendrorhous . Antonie Van Leeuwenhoek 84: 279–287. [DOI] [PubMed] [Google Scholar]
- 56. Niklitschek M, Alcaíno J, Barahona S, Sepúlveda D, Lozano C, Carmona M, et al. (2008) Genomic organization of the structural genes controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous . Biol Res 41: 93–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher’s website.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.