Abstract
Differences in tolerance to water stress may underlie ecological divergence of closely-related ploidy lineages. However, the mechanistic basis of physiological variation governing eco-geographical cytotype segregation is not well understood. Here, using Brachypodium distachyon and its derived allotetraploid B. hybridum as model, we test the hypothesis that, for heteroploid annuals, ecological divergence of polyploids in drier environments is based on trait differentiation enabling drought-escape. We demonstrate that under water limitation allotetraploids maintain higher photosynthesis and stomatal conductance and show earlier flowering than diploids, concordant with a drought-escape strategy to cope with water stress. Increased heterozygosity, greater genetic variability and plasticity of polyploids could confer a superior adaptive capability. Consistent with these predictions, we document (1) greater standing within-population genetic variation in water use efficiency and flowering time in allotetraploids, and (2) the existence of (non-linear) environmental clines in physiology across allotetraploid populations. Increased gas exchange and diminished WUE occurred at the driest end of the gradient, consistent with a drought-escape strategy. Finally, we found that allotetraploids showed weaker genetic correlations than diploids congruous with the expectation of relaxed pleiotropic constraints in polyploids. Our results suggest evolutionary divergence of ecophysiological traits in each ploidy lineage.
Keywords: Polyploidy, hybridization, gas exchange, flowering time, genetic correlations, adaptation
Introduction
Polyploidy (i.e., whole genome duplication) alters DNA content, chromosome number and organization, and gene dosage (Soltis et al. 2004), and has been a pivotal feature of vascular plant diversification (Wood et al. 2009; Jiao et al. 2011). Newly formed polyploid lineages often differ in ecological tolerances and phenotypes from their progenitors (e.g., Chao et al. 2013), which may lead to population divergence and ecological speciation (Levin 2002; Ramsey 2011; Martin and Husband 2013). Polyploidy is a recurrent form of genetic variation (Levin 2002; Parisod 2012), yet we know little about how ploidy influences standing natural variation in ecologically important traits (see however Nuismer and Cunningham 2005; Oswald and Nuismer 2010; Martin and Husband 2012).
In habitats worldwide, water stress influences plant adaptation, abundance, distribution and productivity (Heschel et al. 2002; McKay et al. 2003; Engelbrecht et al. 2007). Plants have evolved several strategies to cope with drought stress, including dehydration avoidance, drought tolerance and escape by reproducing before the onset of drought (McKay et al. 2003; Sherrard and Maherali 2006; Tuberosa 2012). These strategies rely on complex ecophysiological traits whose effects may vary across environments (e.g., Ehrenreich et al. 2009; Xu et al. 2009; Tardieu and Tuberosa 2010). In dry environments, drought-avoiding C3 plants have evolved elevated water-use efficiency (WUE) by closing stomata, which restricts transpiration more than it restricts photosynthesis. This tradeoff between CO2 uptake and water loss depends on stomatal anatomy and conductance, which may be targets of selection (Ackerly et al. 2000; Heschel et al. 2002; Donovan et al. 2007; Agrawal et al. 2008). Alternatively, plants with a drought-escape strategy maintain rapid growth, higher gas exchange, and early flowering, allowing the plant to complete its life cycle before the onset of water limitation (Heschel and Riginos 2005; Sherrard and Maherali 2006; Donovan et al. 2007; Wu et al. 2010). Ecophysiological traits associated with drought tolerance and escape vary genetically within and among populations (Ackerly et al. 2000; Geber and Griffen 2003; Caruso et al. 2005; Culley et al. 2006; Sherrard et al. 2009), which could lead to adaptive population divergence if selection differs across contrasting environments (Ackerly et al. 2000; Donovan et al. 2009). However, adaptive evolution of ecophysiological traits could be constrained if traits are genetically correlated and the sign of the correlations conflicts with the magnitude and direction of selection on the traits (e.g., McKay et al. 2003; Caruso et al. 2005; Sherrard and Maherali 2006); or if expression of genetic covariation depends upon water availability (i.e., genetic tradeoffs across environments; Sherrard et al. 2009; Ivey and Carr 2012).
In heteroploid species, ecological tolerance to water limitation may differ between polyploids and their lower-ploidy ancestors due to morphological and physiological changes derived from polyploidization (Levin 2002; te Beest et al. 2012). In perennial species, polyploid lineages often have greater physiological capability to respond to water deficit than their diploid progenitors through (i) increasing WUE, and reducing stomatal conductance, transpiration and growth rate; and (ii) maximizing water uptake by optimizing plant hydraulic function (Li et al. 1996; Maherali et al. 2009; Allario et al. 2013; Hao et al. 2013). This dehydration-avoidance strategy of polyploids may allow them to differentiate in drier environments than diploids (e.g., Treier et al. 2009; Hao et al. 2013). Alternatively, for heteroploid annuals growing across aridity gradients, ecological differentiation of polyploids in drier environments could be based on a drought-escape strategy involving high gas exchange, low WUE, rapid development and early flowering; this functional strategy is often favored under drought stress in annual species (Heschel and Riginos 2005; Sherrard and Maherali 2006; Donovan et al. 2007; Ivey and Carr 2012). Polyploids that employ a drought-escape strategy would show positive relationships between photosynthesis, stomatal conductance and aridity, and negative relationships between WUE, flowering time and aridity.
The ability of polyploids to colonize new habitats outside of the range(s) of the diploid progenitor may depend on the extent of genetic variation and plasticity in ecophysiology and other key traits (Levin 2002; Hahn et al. 2012; te Beest et al. 2012). Polyploids may show greater genetic variability and increased plasticity compared with diploids because of increased heterozygosity (especially in allopolyploids formed by interspecific hybridization; Doyle et al., 2008), which in turn can confer a higher adaptive response to environmental change (Soltis et al. 2010; Hahn et al. 2012; te Beest et al. 2012; Madlung 2013). Furthermore, increase gene copy number in polyploids could ease pleiotropic constraints (Otto 2007), and enhance the capacity of polyploids to adapt in response to novel selection.
Here, we use the diploid annual grass Brachypodium distachyon and its derived allotetraploid B. hybridum (Poaceae) to examine natural variation and differentiation in ecophysiological traits in diploid (2n =10) and allotetraploid (2n = 30) accessions from multiple wild populations across the Iberian Peninsula. Brachypodium ploidies are geographical and ecologically differentiated (Manzaneda et al. 2012). Both lineages grow in Mediterranean-type climate, which is typically characterized by a relative long estival drought. However, B. distachyon diploids occur frequently in wet habitats where summer drought is attenuated; therefore, a drought-escape strategy might not be adaptive (but see Sherrard and Maherali 2006). In contrast, B. hybridum allotetraploids inhabit mainly dry environments where a predictable summer drought period exists, and a drought-escape strategy could confer fitness advantages, as is true for other winter annual grasses in Mediterranean climates with prolonged dry periods (e.g., Sherrard and Maherali 2006 and references therein). In addition, B. hybridum allotetraploids have larger stomata (Catalán et al. 2012) and are more efficient in their water use than diploids (Manzaneda et al. 2012). Although high WUE is typically associated with a dehydration-avoidance strategy, increased WUE could be advantageous in water-limited environments if it enhances the likelihood of survival to reproduction in a shortened growing period (Wu et al. 2010). Likewise, accessions of this species show significant variation in flowering time (Schwartz et al. 2010), although the adaptive significance of such variation is unknown. The overarching objectives of this study were to evaluate whether physiological differences between Brachypodium ploidies underlie observed ecogeographical differentiation (Manzaneda et al. 2012), and to analyze the potential for adaptive evolution of different Brachypodium ploidy linaeges in populations with contrasting aridity.
By exposing multiple accessions of these lineages to two contrasting watering regimens in the greenhouse, we tested whether Brachypodium ploidies differ in gas exchange physiology and flowering times. We predict that under water limitation, allotetraploids will maintain higher photosynthesis and stomatal conductance and flower earlier than diploids, consistent with a drought-escape strategy. We then analyzed the extent of broad-sense genetic variation within and between populations and plasticity in these traits for each ploidy level, to test the prediction that allotetraploids maintain higher genetic variation and increased plasticity than diploids. We also tested for associations between trait variation and aridity at the level of the population. In particular, we hypothesize that allotetraploid lines from warmer, drier populations will have higher photosynthesis, stomatal conductance and earlier flowering than lines from cooler and wetter populations. Finally, we examined genetic correlations between ecophysiological traits under contrasting water conditions in diploid and allotetraploid lines to identify potential evolutionary constraints to adaptation.
MATERIAL AND METHODS
Study system
B. distachyon (L.) P. Beauv., is a temperate annual grass (10–20 cm) native to the Mediterranean Basin, Middle and Near East (Catalán et al. 2012). This grass inhabits a wide variety of climatic and ecological conditions from sea level to 2000 m elevation; it frequently occurs in forest edges, natural xerophytic meadows, abandoned fields, and along roadsides (Manzaneda et al. 2012). This self-compatible grass (heterozygosity ranges between 0–0.016; Vogel et al., 2009) has a short life-cycle, and germination studies show that both winter-annuals and spring-annuals are present in the Iberian Peninsula (Manzaneda & Rey unpublished). Flowering time varies notably within and between winter-annuals (which require vernalization before flowering) and spring-annuals (which have no vernalization requirement) in response to temperature and light cues (Schwartz et al. 2010). Similarly, B. hybridum allotetraploid genotypes typically do not require vernalization to flower (Martínez & Manzaneda unpublished). Flowering occurs naturally between April-June in the Iberian populations and seed maturation and dispersal occur during June-August.
The B. distachyon complex exhibits variation in somatic chromosome number (2n = 10, 20, 30) (Robertson 1981). Fluorescence in situ hybridization (FISH and GISH) techniques revealed that 2n = 30 is an allotetraploid derived from two diploids (2n = 10 and 2n = 20) (Hasterok et al. 2004), and Catalán et al. (2012) recently proposed that these three cytotypes are in fact three closely-related species: two diploids with a different chromosome base number, B. distachyon (x =5, 2n= 10) and B. stacei (x = 10, 2n= 20), and their derived allotetraploid B. hybridum (x = 5+10, 2n= 30). In the Iberian Peninsula, the two most common cytotypes are the diploid B. distachyon and the allotetraploid B. hybridum whereas populations of B. stacei have been found just in a few localities in SE Spain (Manzaneda et al. 2012; López-Alvarez et al. 2012). B. distachyon diploids (‘diploids’ hereafter) are the most common cytotype in the east and north of the Iberian Peninsula, whereas B. hybridum allotetraploids (‘allotetraploids’ hereafter) are the most frequent cytotype in the south and west of the Iberian Peninsula at low to mid-elevations (Manzaneda et al. 2012). Independent of geography, allotetraploids occur in drier locations than diploids (Manzaneda et al. 2012).
Plant material and experimental procedure
We selected mature seeds from 189 accessions (i.e., 90 diploid and 99 allotetraploid inbred lines derived from our field collection) from 24 populations (~ 8 lines/population) distributed across the Iberian Peninsula (Tables S1, S2 and Fig. S1). Ploidy level of each line was previously determined using flow cytometry and root-tip squashes (Manzaneda et al. 2012). These populations occur in diverse climatic conditions. We chose these populations to maximize natural variation in drought-tolerance traits across a significant part of the range in the study region (Table S1). Nine populations contain uniformly diploid plants, 10 populations contain purely allotetraploids and 5 are mixed populations of diploids and allotetraploids (Tables S1, S2).
In April 2009, we stratified 8 sib seeds of each selfing-inbred line (N =1512 seeds total) at 4 °C for 1 week to facilitate uniform seed germination. We minimized potential maternal effects by using seeds from a first generation of self-fertilized, greenhouse-grown plants. Plants were grown on standard soil (Fafard 4p mix; Fafard Inc., Agawam, MA, USA) at the Duke University greenhouse in 16 h light: 8 h dark at 22 °C in 21 cm ‘conetainers’ (Model SC10R, volume: 164 ml; Stuewe & Sons Inc., Corvallis, OR, USA) in a split-plot design with watering treatment as the whole plot factor. Specifically, plants were exposed in two separate benches (distance between benches was ca. 1 m) to one of two watering treatments: well-watered or water restricted. Each line (189 in total) was replicated four times in separate blocks for each watering treatment (within each block, plants were planted randomly). In the well-watered treatment, plants were kept watered by using bottom watering trays over 9 weeks. In the water-restricted treatment, we simulated natural dry-down conditions experienced by plants in the field after precipitation. Initially, plants were watered to saturation, followed by a 9-week gradual dry-down treatment during which time we watered plants from above (~3.8L/block) just once during the fourth week (Fig. S2). Weekly, we used a volumetric soil moisture probe (TRIME®-PICO-32) to record water content of the soil of three randomly chosen conetainers per block. In the well-watered treatment, soil moisture was always above 65 %, whereas under water-restriction soil moisture averaged 45 ± 6.4 (SE) %, although from the fourth week until the end of the experiment it declined to 20–25 % (ANOVA on soil moisture variation, water treatment x date interaction: F1,8 = 12.63; P = 1.66−10) (Fig. S2). Tissue damage increases due to wilting at soil moisture levels < 20 % , often producing unreliable measurements of gas exchange (i.e., negative gas exchange values and fluctuation of gas exchange curves). For that reason, we set 20 % soil moisture (slightly above the wilting point) as the minimum threshold for obtaining reliable physiological data from our plants. The drought treatment had an effective impact on plant physiology (see Results) and development (Fig. S3).
Ecophysiology and flowering phenology
After 3 weeks of plant growth, we measured physiology of 1161 plants (351 plants died before that point). We quantified whole plant photosynthesis (i.e., carbon gain, A) and stomatal conductance (gs) with an infra-red gas analyzer (IRGA, LI-COR LI-6400 portable photosynthesis system) modified to fit a set of four whole-plant cuvettes in a parallel processing system (see Fig. S4 and Supplementary Materials for methodological details).
We estimated WUE integrated over the lifetime of the leaf from the ratio 13C/12C (carbon isotope composition, δ13C) (e.g., McKay et al. 2003; Sherrard and Maherali 2006; Chen et al. 2011). At the 10th week of the experiment, we collected one leaf per plant for isotope analysis from 95 lines: 48 diploids and 47 allotetraploids, replicated 3 times in each water treatment. Samples were analyzed by isotope ratio mass spectrometry at Duke Environmental Stable Isotope Laboratory (www.biology.duke.edu/jackson/devil).
Because some genotypes require a cold vernalization period to induce flowering (mainly B. distachyon diploid genotypes), after taking gas exchange measurements, we moved plants to a cold room (4°C, 16 h light and 8 h dark, ca. 3 weeks; Fig. S2). To ensure that individuals experienced the same environmental conditions across the experiment, we moved all plants were into the cold room whether they had flowered or not. Plants were then moved back to the greenhouse until the end of the experiment. To quantify flowering time we monitored plants daily once the first plant flowered (after ca. 4 weeks from germination) for ca. 150 days, after which point no remaining plants flowered. We defined flowering time as the number of days from germination until the opening of the first spike.
Statistical analysis
To test differentiation in ecophysiological traits between ploidies and treatments, we conducted general linear mixed models (GLMM) with ploidy, watering treatment and their interaction as fixed effects. We examined the ‘ploidy x treatment’ interaction by conducting tests of simple main effects using the SLICE option in SAS (ver. 9.3, SAS Institute). In these models, we treated line nested in ploidy and block as random effects. To incorporate the split-plot for watering main effect testing, we nested the random block effect within watering treatment. By this way, degrees of freedom for assessing the effect of the watering treatment come from the set of individuals in each block. Because the presence of inflorescences may influence gas exchange (Earley et al. 2009), we included the number of spikes as a covariate in the models.
We analyzed flowering time by fitting a proportional hazards Cox regression model (Proc PHREG, SAS). To account for the effect of differences among lines and blocks in flowering time, line was incorporated as random effect (i.e., ‘frailty’) and block as fixed effect (due to limitations of the statistical package only one random factor can be included in the same model).
We estimated the degree of phenotypic plasticity for each trait and ploidy level from the difference in the cytotype least squares adjusted means between the two watering treatments. Significance of plasticity values comes from interactions slice tests and contrast tests. In addition, for traits with significant plasticity, we analyzed genetic variation in phenotypic plasticity. Due to model convergence issues, we opted to fit separate general linear mixed models to investigate variation in plasticity across lines in each ploidy level. In these models, population was considered a fixed factor, and line nested within population and block as random effects.
To analyze broad-sense genetic variation in ecophysiology within and among populations, we first fit GLMM with restricted maximum likelihood estimates using Proc MIXED, with population as a fixed factor and random effects for line nested within population and block. Statistical inferences (Wald’s Z test) for the covariance parameters for random factors were computed using the COVTEST statement in the procedure MIXED. We fit separate models for each combination of trait, ploidy level and watering regime. Similarly, we obtained the coefficients of variation for each trait. Before and during ecophysiological measurements, 443 plants began flowering. For that reason, we also included the number of spikes produced per each plant at the time of the gas exchange measurements as a covariate in models of whole plant gas exchange variation. Non-flowering individuals (380 plants) were not included in the analyses to estimate genetic variation in flowering time.
We also analyzed the relative contribution of line and population to the total phenotypic variance. We conducted a variance decomposition following a hierarchical design and using restricted maximum likelihood estimates (that account for unbalanced data) in Proc MIXED models that treated population and line (nested within population) as random factors. We compared the fraction of total phenotypic variance for each trait among lines within population (Vline) and among populations (Vpop), with the residual variance (Vres) corresponding to the variance between blocks and individuals within line. We tested whether these variance components differ significantly from zero using a likelihood ratio test with one degree of freedom (e.g., Colautti and Barrett 2011).
For traits with significant genetic variation, we investigated the relationship between trait variation and climate. We used trait population means instead of trait genotypic means to avoid pseudoreplication (see Maron et al. 2007 for a similar procedure). We then analyzed the relationship between population mean trait values and precipitation, temperature and soil moisture deficit separately for each trait and ploidy level. Across our study populations, water availability is not linearly correlated with latitude and longitude (Fig. S5); therefore, we conducted both linear and non-linear regressions. More details on climate correlates are given in Supplementary Materials.
To estimate potential genetic constraints and limits to adaptive evolution for each ploidy lineage, we estimated genetic correlations across individuals of each ploidy level based on variance components from REML (Proc Mixed, SAS) using individual level data between all trait pairs. Genetic correlations computed from a randomly chosen population may be a poor predictor of constraints on adaptive evolution across the range of a species (Sgrò and Hoffmann 2004; Colautti and Barrett 2011). Thus, by including the entire range of trait variation from all populations, we analyzed genetic trait (co)variance in a dataset with maximum range of genetic variation for each trait. Genetic correlations were calculated as:
where Cov (i, j) is the genetic covariance component for traits i and j, and σi and σj are the square roots of the among genotype variance components for traits i and j (e.g., McKay et al. 2003). Significance was determined by using log-likelihood ratios tests comparing full models with reduced models in which the covariance was constrained to 0. We used the full data set to estimate correlations among whole plant gas exchange traits and flowering time on all plants. We used the subset of individuals for which δ13C had been determined to estimate correlations between this trait and the rest of traits. Because genetic correlations may differ across environments (Sgrò and Hoffmann 2004; Sherrad et al. 2009), all correlations were computed from both treatments. Finally, we employed Cheverud’s random skewers method (Cheverud and Marroig 2007) to quantitatively compare genetic variance-covariance matrices between ploidy lineages within treatment, or between treatments within each ploidy lineage. If the mean response cosine is larger than 95% of the cosine of random vectors, then we cannot reject the null hypothesis that the two G-matrices are equal or proportional. See Supplementary Materials for further details on this procedure.
For all analyses, photosynthesis and stomatal conductance were log-transformed to improve normality and homoscedasticity.
RESULTS
Trait differentiation
Ecophysiological traits
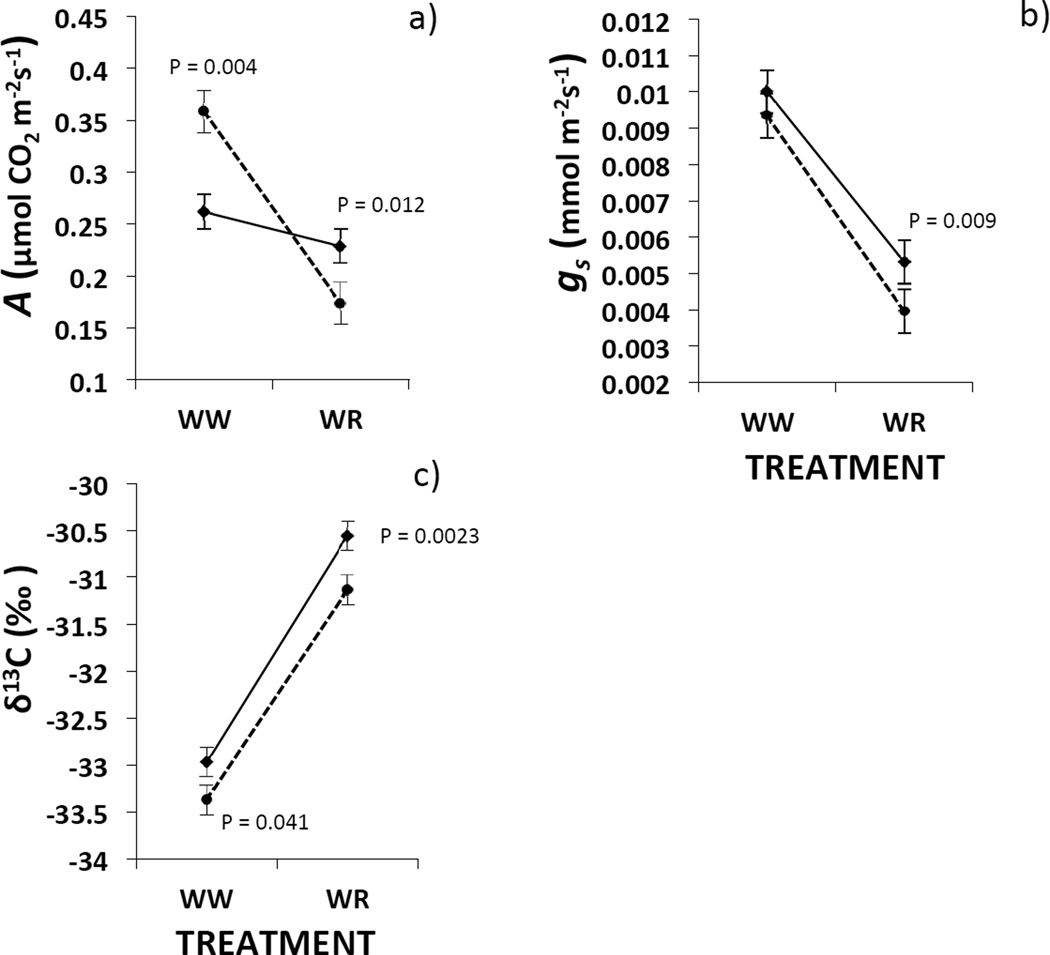
The effect of watering on photosynthesis differed for each ploidy level (i.e., significant ploidy x treatment; Table 1). Under well-watered conditions, diploids showed significantly higher whole plant carbon gain than allotetraploids (Fig. 1A). In contrast, under water-restricted conditions, diploids had significantly lower carbon gain than allotetraploids (Fig 1A). Ploidy significantly affected stomatal conductance and δ13C (Table 1). Allotetraploids had higher stomatal conductance and WUE (higher δ13C) than diploids (Fig. 1B,C), especially under water-restricted treatment (Fig. 1B,C). In all traits, there was significant among-line variation within each ploidy level (Table 1). Finally, the number of spikes was positively associated with whole carbon gain and stomatal conductance (0.035±0.005, 0.065±0.005, b±1SE for A and gs respectively).
Table 1.
Summary results of general lineal mixed models testing the effects of ploidy (P), water treament (T) and their interaction on variation of ecophysiological traits across multiple B. distachyon diploid and B. hybridum allotetraploid lines. The effect of ‘Line’ nested within ploidy was incorporated as a random factor in the models. We included the number of spikes as a covariate in gas exchange models. Significant values (P < 0.05) are in bold.
| A (µmol CO2 m−2s−1) | gs (mmol m−2s−1) | Integrated WUE (δ13C) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Source | d.f. | F | P | d.f. | F | P | d.f. | F | P |
| Ploidy (P) | 1, 169 | 0.08 | 0.78 | 1, 165 | 8.79 | 0.0035 | 1, 93 | 8.15 | 0.0053 |
| Water treatment (T) | 1, 6.21 | 7.21 | 0.0351 | 1, 6.24 | 73.98 | 0.0001 | 1, 4.28 | 182.72 | 0.0001 |
| P x T | 1, 981 | 36.84 | <0.0001 | 1, 977 | 2.63 | 0.105 | 1, 414 | 1.13 | 0.289 |
| Random Effects | Z | P | Z | P | Z | P | |||
| Line(Ploidy) | 5.38 | <0.0001 | 4.89 | <0.0001 | 4.86 | <0.0001 | |||
| Covariate | d.f. | F | P | d.f. | F | P | d.f. | F | P |
| Number of spikes | 1, 1006 | 78.93 | <0.0001 | 1, 876 | 125.77 | <0.0001 | NA | NA | NA |
FIGURE 1.
Variation (LS-MEANS adjusted model values ± 1 SE) in a) whole plant photosynthesis (A); b) stomatal conductance (gs); c) carbon isotope composition (δ13C ‰) between two watering treatments (WW = well watered, WR = water restricted) and two levels of ploidy variation: B.distachyon diploids (dashed line) and B. hybridum allotetraploids (solid line). P-values in the graphs are results from test of the slices interactions comparing both ploidy levels in each treatment. Only results from significant tests are shown.
Flowering phenology
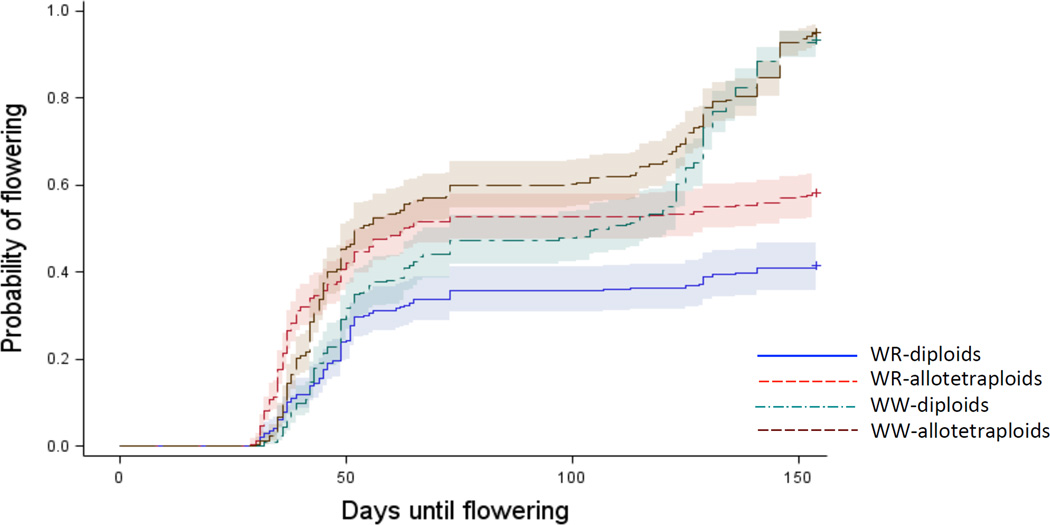
During the experiment 70.9 % of plants (N = 1307) flowered. The effect of water level depended on ploidy (ploidy x treatment likelihood ratio test: χ2 = 5.77, df = 1, P = 0.016; treatment: χ2 = 8.43, df = 1, P = 0.0036; ploidy χ2 = 0.322, df = 1, P = 0.57). Under well-watered conditions, diploids and allotetraploids had essentially the same probability of flowering through time (df = 1, χ2 = 2.05, P = 0.152; Fig. 2). However, under water restriction diploids had a 34.6% lower probability of flowering than allotetraploids (df = 1, χ2 = 7.75, P = 0.0054; Hazard Ratio 0.654, 95% CL: 0.528–0.812; Fig. 2).
FIGURE 2.
The probability of flowering for B.distachyon diploid and B. hybridum allotetraploid individuals in each watering treatment as a function of time (number of days). Shading indicates 95 % confidence intervals.
Phenotypic plasticity
Both diploids and allotetraploids showed significant plasticity to watering treatment for all traits analyzed (Table S3; Fig. 1). Plasticity varied between ploidies only for carbon gain, with diploids showing higher plasticicty in this trait (ANOVA: F1,164 = 13.17, P = 0.0004; Table S3).
For diploids, there was significant genetic variation in plasticity among populations in all traits (Table 2). In addition, gs plasticity also varied among lines within population (Table 2). Plasticity of allotetraploids significantly differed at population level for gs and flowering time (Table 2).
Table 2.
Summary results of the general linear mixed analyses conducted to test for genetic variation at population and family level (i.e., significance of factor ‘line’) in plasticity of physiological traits and flowering time across B. distachyon diploid and B. hybridum allotetraploid lines.
| Diploids | Population |
Line (Population) |
|||
|---|---|---|---|---|---|
| Traits | DF | F | P | Z | P |
| A (µmol CO2 m−2s−1) | 13,77 | 2.26 | 0.014 | 0.45 | 0.651 |
| gs (mmol m−2s1) | 13,59 | 8.77 | <0.0001 | 1.99 | 0.046 |
| Integrated WUE (δ13C ‰) | 13,33 | 3.64 | 0.001 | 0.77 | 0.44 |
| Flowering time (n° days) | 13,79 | 2.2 | 0.016 | 1.07 | 0.285 |
| Allotetraploids | |||||
| A (µmol CO2 m−2s−1) | 14,72 | 0.51 | 0.917 | 1.03 | 0.301 |
| gs (mmol m−2s1) | 14,87 | 2.31 | 0.0095 | 0.2 | 0.838 |
| Integrated WUE (δ13C ‰) | 14,34 | 1.17 | 0.334 | 1.34 | 0.181 |
| Flowering time (n° days) | 14,65 | 2.06 | 0.026 | 0.48 | 0.633 |
Extent of genetic variation
We found extensive genetic variation in physiology and flowering time, although in most cases population rather than lines within population accounted for the genetic variation in these traits (Table 3; Figures S6, S7). With the exception of gs, trait variation was higher under water restriction than in well-watered conditions (Table 3). Under well-watered conditions, allotetraploids showed higher trait variation than diploids in photosynthesis and flowering time, similar variation in δ13C, and lower variation in gs. Under water-restriction, allotetraploids showed higher levels of variation than diploids in flowering time, δ13C, similar in gs, and lower in photosynthesis (Table 3). In both ploidies, the number of spikes was positively associated with whole carbon gain and gs (Table S4) under both watering treatments.
Table 3.
Coefficient of variation of physiological traits and flowering time in B. distachyon diploid (within each cell, data above = D) and B. hybridum allotetraploid (within each cell, data below = T) lines under two watering regimes. A summary of the results of the general or generalized linear mixed analyses conducted to test for broad-sense genetic variation within and between populations in these traits is also shown. Significant values (P < 0.05) are in bold.
| Well watered | ||||||
|---|---|---|---|---|---|---|
| Source | Line (Population) | Population | ||||
| Trait | Coefficient of variation (%) |
Z | P | DF | F | P |
| A (µmol CO2 m−2s−1) | D: 69.19 T: 73.64 |
0.02 1.01 |
0.983 0.312 |
13,87 14,79 |
5.19 3.92 |
<0.0001 <0.0001 |
| gs (mmol m−2s1) | D: 152.85 T: 131.50 |
0.6 0.24 |
0.549 0.814 |
13,83 14.77 |
7.3 6.19 |
<0.0001 <0.0001 |
| Integrated WUE (δ13C ‰) | D: 3.34 T: 3.26 |
0.07 1.88 |
0.946 0.029 |
13,28 14,31 |
8.97 1.40 |
<0.0001 0.211 |
| Flowering time (# of days) |
D: 48.29 T: 54.66 |
5.0 4.21 |
<0.0001 <0.0001 |
13,74 14,81 |
19.87 17.85 |
<0.0001 <0.0001 |
| Water restricted | ||||||
| A (µmol CO2 m−2s−1) | D: 107.6 T: 98.6 |
1.11 0.87 |
0.268 0.384 |
13,75 14,66 |
2.27 4.34 |
0.014 <0.0001 |
| gs (mmol m−2s1) | D: 125.79 T: 125.33 |
0.31 0.47 |
0.758 0.635 |
13,72 14,72 |
3.55 6.11 |
0.0003 <0.0001 |
| Integrated WUE (δ13C ‰) | D: 4.01 T: 4.34 |
0.97 1.74 |
0.333 0.041 |
13,33 14,34 |
7.80 2.25 |
<0.0001 0.027 |
| Flowering time (# of days) |
D: 54.54 T: 58.74 |
2.93 3.62 |
0.0034 0.0003 |
12,40 13,48 |
5.53 2.94 |
<0.0001 0.0032 |
For diploids, trait variance was greater among populations than among lines within population in both treatments (Table 4). For allotetraploids, whole carbon gain, gs and flowering time exhibited significantly greater phenotypic variance among populations than genetic variance within populations, but the opposite was true for δ13C in both treatments, and for flowering time under water restriction (Table 4). For δ13C variance among lines was higher in allotetraploids than in diploids in both treatments (Table 4), as was flowering time under water restriction (Table 4). Among-population fraction of phenotypic variance was slightly higher for diploids than for allotetraploids in the well-watered treatment. Under water restriction, such fraction was higher for diploids than for allotetraploids for δ13C and flowering time but lower for carbon gain and gs (Table 4).
Table 4.
Fraction of total phenotypic variance of physiological drought tolerance-related traits (and flowering time) accounted for within and among populations in B. distachyon (diploid) and B. hybridum (allotetraploid) accessions sampled across the Iberian Peninsula and grown under two different watering treatments. Variance components are standardized to sum to one. Variance components significantly different from zero (P < 0.05) are in bold.
| Well watered | Diploids | Allotetraploids | ||||
|---|---|---|---|---|---|---|
| Traits | Vline | Vpop | Vres | Vline | Vpop | Vres |
| A (µmol CO2 m−2s−1) | 0 | 0.194 | 0.805 | 0.085 | 0.189 | 0.725 |
| gs (mmol m−2s1) | 0.041 | 0.328 | 0.631 | 0.106 | 0.281 | 0.642 |
| Integrated WUE (δ13C ‰) | 0 | 0.468 | 0.532 | 0.279 | 0.086 | 0.625 |
| Flowering time (days) | 0.177 | 0.704 | 0.118 | 0.164 | 0.603 | 0.231 |
| Water restricted | ||||||
| A (µmol CO2 m−2s−1) | 0.107 | 0.125 | 0.768 | 0.091 | 0.203 | 0.706 |
| gs (mmol m−2s1) | 0.021 | 0.13* | 0.848 | 0 | 0.19 | 0.81 |
| Integrated WUE (δ13C ‰) | 0 | 0.347 | 0.653 | 0.187 | 0.143 | 0.67 |
| Flowering time (n° days) | 0.247 | 0.478 | 0.274 | 0.473 | 0.213 | 0.313 |
P<0.1
Climatic correlates
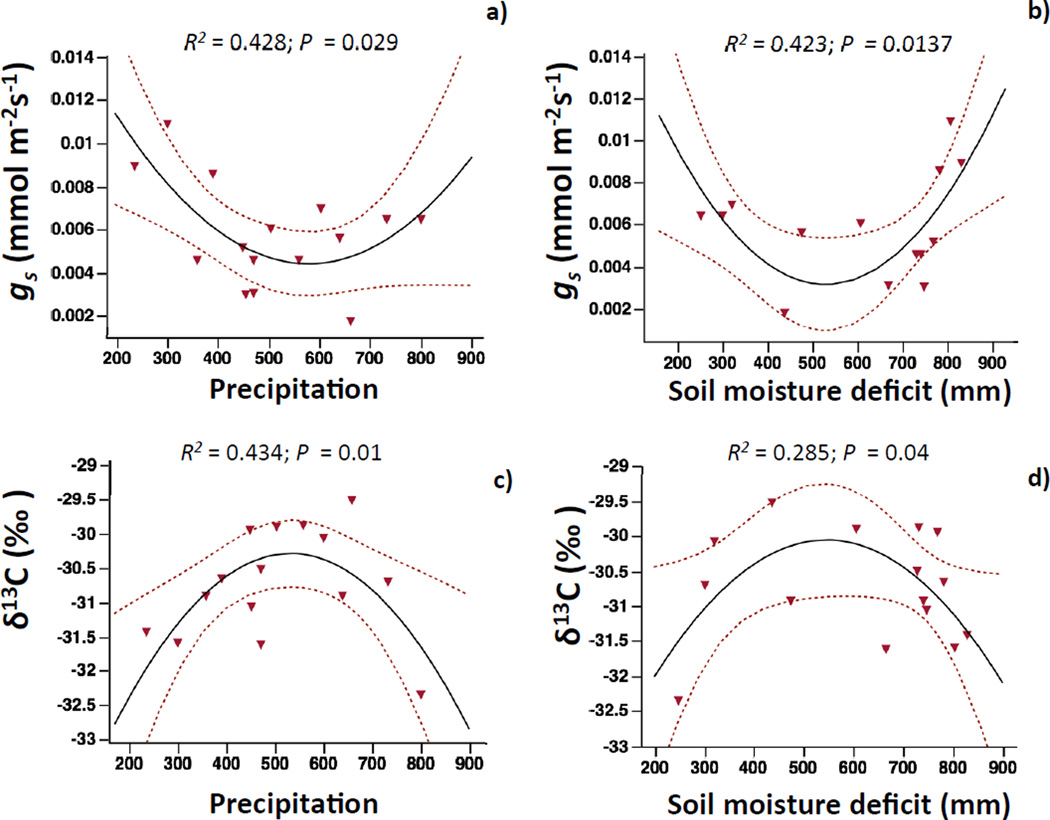
We found significant associations between trait variation and climate only across allotetraploid populations. Relationships of gs and δ13C with climatic factors were non-linear (Fig. 3A–D). Populations with low or high precipitation and soil moisture deficit had higher gs, whereas intermediate climates had minimal stomatal conductance (Fig. 3A,B). A similar trend was also observed for photosynthesis and soil moisture deficit, although significance was marginal (P<0.1; Fig. S8B). δ13C was maximum in populations with mid-values of precipitation and soil moisture deficit and decreased in populations with higher or lower aridity (Fig. 3C,D). Genotypic variation in photosynthesis correlated positively with temperature and negatively with precipitation, although significance was marginal (P<0.1; Fig. S8C,D). Flowering time correlated positively with precipitation although significance was also marginal (P<0.1; Fig. S8E).
FIGURE 3.
Relationships between annual precipitation and annual soil moisture deficit and the genetic population means of ecophysiological traits under water-restricted conditions: a,b) stomatal conductance (gs); c,d) carbon isotope composition (δ13C ‰). Soil moisture deficit is defined as potential evapotranspiration minus precipitation. In all cases, the solid line depicts the best quadratic regression fit between variables, and dashed lines are its 95% confidence intervals. Only data for B. hybridum allotetraploids are depicted (red triangles).
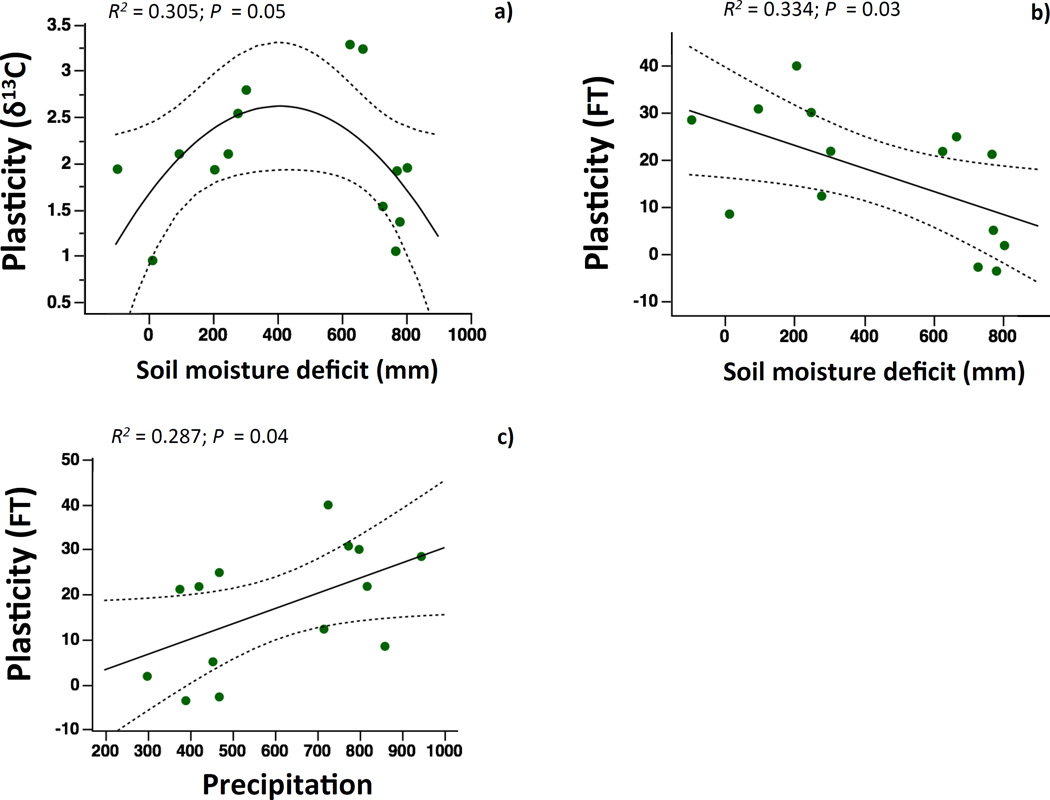
We found significant associations between trait plasticity and climate only across diploids populations. The relationship between δ13C plasticity and soil moisture deficit was non-linear (Fig 4A). δ13C plasticity was maximum in populations with intermediate values of soil moisture deficit and decreased in populations with higher or lower aridity. Plasticity in flowering time increased with precipitation and decreased with soil moisture deficit (Fig. 4B,C). We detected no significant climatic relationships between climate and the remaining genetically-variable traits (Table 2).
FIGURE 4.
Relationships between annual precipitation, annual soil moisture deficit and population means of plasticity in ecophysiological traits: a) carbon isotope composition; b,c) flowering time (FT). Soil moisture deficit is defined as potential evapotranspiration minus precipitation. Plasicity values come from trait differences between the two watering treatments. In all cases, solid lines depict the best linear or quadratic regression fit between variables. Only data for B. distachyon diploids are depicted by green circles in the graphs.
Genetic correlations
For diploids, all traits were significantly genetically correlated with each other in both treatments (Table 5). We found positive genetic correlations between photosynthesis and gs, and between δ13C and flowering time in both treatments, and negative correlations for the remaining traits (Table 5). For allotetraploids, the number of significant genetic trait correlations was overall lower. We found a significant positive genetic correlation between photosynthesis and gs under water restriction (Table 5). Flowering time was negatively correlated with photosynthesis and gs in both treatments (Table 5). For both ploidy levels, the sign of genetic correlations never changed across treatments.
Table 5.
Genetic correlations ± SE between 4 ecophysiological traits for B. distachyon diploids (above the diagonal, WR: N = 90 lines, N = 47 lines for δ13C correlations; WW: N = 89 genotypes, N = 47 genotypes for δ13C correlations), and B. hybridum allotetraploids (below the diagonal, WR: N = 98 lines, N = 47 lines for δ13C correlations; WW: N = 94 genotypes, N = 46 genotypes for δ13C correlations) in the two water treatments. Genetic correlations were estimated from variance components in REML (Proc Mixed) using individual level data. Traits were standardized to a mean of 0 and standard deviation of 1. Uncorrected P-values are presented beneath the genetic correlations. Significant genetic correlations after Bonferroni correction for 6 tests for each ploidy level (α = 0.05/6=0.008) are in bold.
| Water restricted (WR) |
A | gs | δ13C | Flowering time |
|---|---|---|---|---|
| A |
1.0±0.023 P<0.0001 |
0.999± 0.21 P=0.0003 |
−0.72± 0.10 P<0.0001 |
|
| gs |
0.95 ± 0.072 P=0.0001 |
− |
−0.79± 0.103 P<0.0001 |
|
| δ13C | 0.60 ±0.34 P=0.107 |
0.62 ± 0.65 P=0.32 |
−0.83 ± 0.13 P<0.0001 |
|
| Flowering time |
−0.89 ± 0.10 P<0.0001 |
−0.86 ± 0.12 P<0.0001 |
−0.53 ± 0.19 P=0.0121 |
|
| Well-watered (WW) |
A | gs | δ13C | Flowering time |
| A |
0.98±0.043 P<0.0001 |
0.86±0.14 P<0.0001 |
−0.81 ±0.097 P<0.0001 |
|
| gs | − |
0.62±0.15 P=0.0013 |
−0.66±0.094 P<0.0001 |
|
| δ13C | 0.20±0.31 P=0.53 |
0.45±0.74 P=0.53 |
−0.74±0.11 P<0.0001 |
|
| Flowering time |
−0.71 ±0.105 P<0.0001 |
−0.80±0.14 P<0.0001 |
−0.35±0.21 P=0.12 |
|
(−) Non-estimable because model fails to converge.
The analyses of random skewers showed that response vectors of G-matrices between ploidy lineages were not proportional under water restriction (vector correlation = 0.81, P = 0.948), yet they were in the well-watered treatment (vector correlation = 0.85, P = 0.962). In this last treatment, however, response vectors were larger in allotetraploids (response vector length ratio diploid/allotetraploid = 0.81), suggesting that the response to selection may be weaker in diploids. Response vectors differed between treatments in diploids (vector correlation = 0.80, P = 0.947), but not in allotetraploids (vector correlation = 0.87, P = 0.968). The response to selection of allotetraploids may be stronger under well-watered conditions (response vector length ratio water-restricted/well-watered = 0.72).
DISCUSSION
Until recently, the mechanistic basis of physiological variation underlying differential cytotype distribution was not well understood (but see Buggs and Pannell 2007; Hao et al. 2013). Here, we demonstrate that Brachypodium diploids and polyploids differ in gas exchange physiology and flowering time under water limitation. In dry conditions, allotetraploids maintain higher photosynthesis and stomatal conductance, and flower earlier, than diploids, concordant with a drought-escape strategy. For allotetraploids, ecophysiology varied as a function of precipitation and aridity of the provenance populations. Furthermore, consistent with our expectations, adaptive evolution of ecophysiology is likely constrained by low levels of genetic variability and the presence of genetic correlations, in particular for B. distachyon diploid genotypes occurring naturally in wet habitats. In addition, we found extensive genetic trait differentiation among Iberian Brachypodium populations. Consistent with predictions, standing within-population genetic variation in integrated WUE and flowering phenology was greater in allotetraploids than diploids. Overall, our study indicates that the divergence in ecological tolerance to water stress between Brachypodium ploidy lineages is likely an important driver of ecogeographical differentiation in this species complex (Manzaneda et al. 2012).
Adaptive significance of trait differentiation and plasticity
Here, we show that trait differentiation between Brachypodium ploidies depends on environment for photosynthesis, flowering time, and to some extent, stomatal conductance, whereas differentiation in integrated WUE was essentially genetic. The elevated photosynthetic performance of diploids under well-watered conditions, suggests that B. distachyon diploids are better adapted than allotetraploids to humid environments. In contrast, allotetraploids maintain higher gas exchange rates, WUE, and accelerated flowering times in dry conditions, suggesting that B. hybridum allotetraploids might be better adapted – through a drought-escape strategy – than diploids to dry environments. Indeed, B. distachyon diploids occur mainly in humid locations, whereas B. hybridum allotetraploids occupy drier sites (Manzaneda et al. 2012). Although decreased WUE is also expected under water stress for short-lived annuals with a drought-escape strategy (as result of increasing stomatal conductance to increase carbon gain; e.g., McKay et al. 2003; Heschel and Riginos 2005), increased WUE could be advantageous in water-limited environments if reduces development time and enhances the likelihood of survival to reproduction in short growing period (Wu et al. 2010). Indeed, under water stress, allotetraploids not only flowered significantly earlier than diploids, but they had ca. 65 % higher probability of flowering. We found negative genetic correlations between flowering time and physiological traits for allotetraploids under water stress, indicating that genetic lines with higher photosynthesis and stomatal conductance flower early (Fig. S9). Negative, genetically-based correlations between gas exchange physiology and flowering time have been reported previously in annuals species and have been interpreted as evidence of drought escape (e.g., Geber and Dawson 1997; McKay et al. 2003; Heschel and Riginos 2005; Sherrard and Maherali 2006; Donovan et al. 2007; Wu et al. 2010).
Allotetraploids maintained higher integrated WUE in dry conditions than diploids, likely owing to a significant reduction in stomatal conductance, but no change in photosynthesis under water stress. Therefore, ecophysiological variation between ploidies under water restriction may reflect cytotypic differences in stomatal anatomy and conductance, as well as differences in photosynthetic capacity (Geber and Dawson 1997; McKay et al. 2003). Indeed, whole genome duplications can have large effects on plant anatomy and gas exchange biochemistry (Warner and Edwards 1993; te Beest et al. 2012). In the case of allopolyploids, however, characteristics of naturally occurring polyploid plants represent not only the inherent effects of increased chromosome number, but also past interspecific hybridization and the results of natural selection after polyploidization (Otto 2007; Maherali et al. 2009). B. hybridum allotetraploids have a polytopic origin resulted from ancient bi-directional crosses of B. distachyon and B. stacei plants acting either as maternal or paternal parents (López-Alvarez et al. 2012). Environmental niche data have shown that B. distachyon occurs in higher, cooler and wetter places than B. stacei, which grows in lower, warmer and drier environments (López-Alvarez et al. in review). Although B. hybridum may also occurs in zones with intermediate climates it frequently grows in low altitudinal warmer and drier places, like its B. stacei progenitor (Manzaneda et al. 2012; López-Alvarez et al. 2012, in review). In fact, across its native range of distribution sympatric admixed populations of B. stacei – B. hybridum have been found in several localities (López-Alvarez et al. in review; A.J. Manzaneda pers. obs). Interestingly, both species share common life-history traits as rapid plant development, lack of vernalization and early flowering (Iberian B. stacei lines bloom ca. 31–39 days after germination; Martínez and Manzaneda unpublished), which are typical features associated to drought-escape. Our results contrast with the only other study of adaptive ecophysiological differentiation in heteroploid annual species, which found that diploid Mercurialis annua are more drought tolerant than polyploids (Buggs and Pannell 2007). Polyploid Mercurialis annua showed lower WUE and photosynthesis and higher transpiration than diploids under water limitation (Buggs and Pannell 2007). Physiological responses to water stress of cytotypes are likely species-specific or could occur via evolutionary changes after polyploidization (Maherali et al. 2009). More studies of heteroploid annuals are needed to evaluate what functional strategy drives adaptive differentiation of polyploids in drier environments.
Fitness and trait data from field studies are necessary to demonstrate whether WUE and gas exchange traits are adaptive. However, adaptive differentiation of ecophysiological traits may be also inferred from the analysis of clinal variation across environmental gradients (e.g., Maron et al. 2007; Paccard et al. 2014). Our correlations between physiology and climate provides additional evidence for adaptive trait differentiation of allotetraploids. First, we detected a significant correspondence between physiological trait variation and climate only across allotetraploid populations. Consistent with a drought-escape strategy, allotetraploid lines from warmer and/or drier populations tended, overall, to have higher photosynthesis and earlier flowering than lines from cooler and/or wetter populations (Fig. S8). However, the strongest relationships between plant functional traits and aridity were not linear. Thus, stomatal conductance and photosynthesis were lowest at intermediate climates and enhanced at both extremes of the aridity gradient. Likewise, WUE peaked in lines from intermediate levels of aridity or annual precipitation. In C3 plants WUE is expected to increase with aridity (Prentice et al. 2011; Cernusak et al. 2013). The non-linear relationships likely reflects the non-linear soil water availability across allotetraploids populations (Fig. S5). When aridity is low to moderate sides of the gradient, increasing aridity is correlated with reduced stomatal conductance and photosynthesis and enhanced WUE, characteristic of a typical dehydration-avoidance response. However, at the driest end of the aridity gradient where selection pressure exerted by water stress is presumably strongest, increasing aridity corresponds with increased stomatal conductance and photosynthesis and diminished WUE, consistent with a drought-escape strategy. Adoption of mixed strategies of resistance and escape in response to drought has been documented previously (e.g., Paccard et al. 2014). Our results suggest mixed strategies across B. hybridum populations that could emerge as result of a differential adaptation to non-linear variations in water availability across the species range. Clinal differentiation among populations in ecophysiology indicates that B. hybridum allotetraploids may have adapted to differences in aridity along climatic gradients, likely resulting from divergent selection exerted by water availability (Ackerly et al. 2000; Donovan et al. 2009). In contrast, trait differentiation among diploid populations may not be adaptive, as we found no relationship between trait variation and climate in diploids. Population differentiation in diploids could be caused by historical or demographical factors, and maintained by limited gene flow between populations (B. distachyon is strongly selfing; Vogel et al. 2009). Reciprocal transplant studies will be required to test whether population differentiation is adaptive.
Many plant species show plasticity in ecophysiological traits when exposed to environments with contrasting water availability (e.g., Agrawal et al. 2008; Ivey and Carr 2012; reviewed in Nicotra and Davidson 2010). We predicted that polyploids would maintain greater plasticity than diploids because of higher levels of heterozygosity and novel genetic and genomic rearrangements derived from polyploidization (te Beest et al. 2012; Hahn et al. 2012). However, we did not detect significantly higher trait plasticity in allotetraploids compared to diploids. In contrast, plasticity in photosynthesis was higher in diploids than allotetraploids. Few studies have compared plasticity among related ploidy lineages, and overall patterns remain ambiguous and trait-dependent (e.g., Bretagnolle and Thompson 2001; Hahn et al. 2012; te Beest et al. 2012). Only for B. distachyon diploids was plasticity related to climate. For diploids, plasticity in WUE varied non-linearly with aridity. The highest values of WUE plasticity were found for populations with intermediate levels of aridity. Plasticity in flowering time varied linearly with aridity/precipitation; higher plasticity in flowering time was found for humid populations. If plasticity in WUE and flowering time is adaptive at mild and wet climates, it could have facilitated cytotypic differentiation and establishment of B. distachyon diploids in such environments.
Limitations on adaptive evolution of drought-response traits in Brachypodium
In our study, ecophysiological traits varied among Brachypodium lines and populations. Previous studies have documented significant natural variation in leaf water content and photochemical efficiency (Luo et al. 2011), flowering and vernalization time (Schwartz et al. 2010) and root system architecture (Pacheco-Villalobos and Hardtke 2012) among B. distachyon accessions from Turkey and Iraq. Those results, and ours from Iberian populations, demonstrate extensive natural variation in drought-response traits across the native range of B. distachyon. Yet, little is known about the adaptive significance of this variation (Pacheco-Villalobos and Hardtke 2012; IBI 2014).
We predicted that allotetraploids would have more genetic variation in ecophysiology than diploids because of increased heterozygosity. As expected, we detected greater genetic variation within populations of allotetraploids than diploids in δ13C in both experimental conditions, and in flowering time under water limitation. Both allotetraploids and diploids maintained low levels of standing genetic variation in the remaining traits, which could limit their adaptive evolution (Caruso et al. 2005). We predict that response to natural selection on δ13C and flowering time should be faster in allotetraploid than in diploid lines, although conclusions about the limits of adaptive evolution in diploids should be taken with caution given the relative low number of lines per population included in this study. Other studies have also reported low heritability of ecophysiological traits associated with drought response (e.g., Heschel et al. 2002; Sherrard and Maherali 2006; Donovan et al. 2007, 2009), and may reflect strong directional selection in the past (Ackerly et al. 2000; see also Geber and Griffen 2003), or the existence of a strong environmental component to measuring gas exchange, which might also reduce the ability to detect additive genetic variation (e.g., Ackerly et al. 2000).
Empirical studies comparing response to selection between ploidy lineages are scarce and it is still unclear whether genome duplication typically increases or decreases the species’ ability to respond to selection (see Martin and Husband 2012). However, our results are consistent with studies of Brassica napus that have documented extensive de novo variation in life-history traits, including flowering time, in new-formed allopolyploid lines compared to their diploid parents (Schranz and Osborn 2004). For both ploidies, levels of genetic variation in ecophysiological traits were significantly higher among populations than within populations indicating substantial population differentiation in physiology. In well-watered conditions, with the exception of δ13C, population-level phenotypic variance was only slightly higher in diploids than allotetraploids. In dry conditions, this pattern was more unpredictable and trait-dependent, but for δ13C, which the amount of phenotypic variance among populations was clearly higher (ca. 36.6%) in diploids than in allotetraploids.
Consistent with our predictions, we found that allotetraploids show weaker genetic correlations than diploids. For diploids, all traits were significantly correlated with each other in both treatments, whereas for allotetraploids the number of significant genetic correlations was lower. Likewise, genetic covariances tend to constrain the responses to selection vectors in similar directions but to varying degrees in both ploidy lineages. Under water-restriction, G-matrices were not entirely proportional between ploidy lineages suggesting different trajectories in the evolution of ecophysiological traits in dry conditions. In wet conditions, although G-matrices were proportional between ploidy lineages, the response vector differed in magnitude suggesting a weaker response to selection of diploids, which is likely caused by a stronger trait correlational structure. As the evolutionary response to natural selection may be constrained by the genetic-covariance matrix, changes in G-matrices associated with polyploidy and/or ancient hybridization could allow allotetraploids to diversify rapidly even if diploid and allotetraploids experience similar patterns of selection (Oswald and Nuismer 2010).
Since diploids occur primarily in mesic populations, we hypothesize that evolution of ecophysiological traits should be constrained across diploid lines and populations because of the existence of strong genetic correlations in humid conditions, consistent with the lack of clinal trait differentiation observed across diploid populations. We are not aware of any study comparing G-matrices of diploid and allopolyploid lineages, but these results are expected from the fact that epigenetic changes, changes in gene expression and chromosomal rearrangements are typically observed in allopolyploids (Oswald and Nuismer 2010; Martin and Husband 2012) which may underlie the weaker genetic correlations observed for B. hybridum allotetraploids. Similarly, proportionality or magnitude of G-matrices differed between watering treatments. For diploids, response vectors of G-matrices were not proportional between watering treatments. For allotetraploids, response vectors differed in magnitude, suggesting that the response to selection may be stronger in wet conditions. Our results are concordant with other studies that reported variations in physiological G-matrices across watering environments (e.g., Sherrard et al. 2009).
Finally, we found pairwise negative genetic correlations between gas exchange physiology, δ13C and flowering time, which could indicate pleiotropy, and might reflect tradeoffs between different drought strategies (e.g., McKay et al. 2003; Heschel and Riginos 2005; Sherrard and Maherali 2006; Donovan et al. 2007). As expected, we found positive genetic correlations among gas exchange traits, which likely reflect tradeoffs between carbon fixation and water loss through the stomata (Geber and Dawson 1997). Both negative and positive genetic correlations could hinder adaptation if the direction of selection is the same for negatively correlated traits, or different for positively correlated traits (Etterson and Shaw 2001; Caruso et al. 2005). Whether such trait genetic correlations are in the same direction as selection (e.g., correlational selection) or opposed to selection still would require experimental verification in natural populations of both ploidy lineages.
Conclusion
Our results support the hypothesis that differences in tolerance to water stress may underlie ecological divergence of closely-related ploidy lineages growing under contrasting environments (Ramsey 2011; Martin and Husband 2013). Adaptive evolution of ecophysiological traits could be constrained by the absence of sufficient trait genetic variation and the structure of genetic correlations between traits. Standing genetic variation and clinal variation of traits differed between ploidies and water environments, suggesting that response to selection of some traits, and their evolutionary trajectories, may be different for each ploidy lineage.
Supplementary Material
Acknowledgments
We thank four anonymous reviewers for valuable comments on the manuscript. We thank J.M. Alcántara for advice on matrix comparisons. We thank Kathy Ghattas and the Duke Greenhouse staff for technical assistance. We are also in debt to Jon Karr and the Duke Environmental Stable Isotope Laboratory staff for the isotope analyses. This project was funded by the European Commission through a Marie Curie Action awarded to AJM and PJR (PIOF-GA-2008-220983) and by the Spanish Ministry of Economy and Competitiveness and FEDER funds from the EC to AJM (RYC-2010-06237; CGL2012-30838). TMO was funded by the National Science Foundation (award EF-0723447) and the National Institutes of Health (award R01 GM086496).
REFERENCES
- Ackerly DD, Dudley S, Sultan SE, Schmitt J, Coleman JS, Linder CR, Sandquist DR, Geber M, Evans AS, Dawson TE, Lechowicz MJ. The evolution of plant ecophysiological traits: recent advances and future directions. BioScience. 2000;50:979–995. [Google Scholar]
- Agrawal AA, Erwin AC, Cook SC. Natural selection on and predicted responses of ecophysiological traits of swamp milkweed (Asclepias incarnata) J. Ecol. 2008;96:536–542. [Google Scholar]
- Allario T, Brumos J, Colmenero-Flores JM, Iglesias DJ, Pina JA, Navarro L, Talon M, Ollitrault P, Morillon R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2013;36:856–868. doi: 10.1111/pce.12021. [DOI] [PubMed] [Google Scholar]
- Bretagnolle F, Thompson JD. Phenotypic plasticity in sympatric diploid and autotetraploid Dactylis glomerata . Int. J. Plant Sci. 2001;162:309–316. [Google Scholar]
- Buggs RJA, Pannell JR. Ecological differentiation and diploid superiority across a moving ploidy contact zone. Evolution. 2007;61:125–140. doi: 10.1111/j.1558-5646.2007.00010.x. [DOI] [PubMed] [Google Scholar]
- Caruso CM, Maherali H, Mikulyuk A, Carlson K, Jackson RB. Genetic variance and covariance for physiological traits in Lobelia: are there constraints on adaptive evolution? Evolution. 2005;59:826–837. [PubMed] [Google Scholar]
- Catalán P, Müller J, Hasterok R, Jenkins G, Mur LAJ, Langdon T, Betekhtin A, Siwinska D, Pimentel M, López-Alvarez D. Evolution and taxonomic split of the model grass Brachypodium distachyon . Ann. Bot-London. 2012;109:385–405. doi: 10.1093/aob/mcr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernusak LA, Ubierna N, Winter K, Holtum JAM, Marshall JD, Farquhar GD. Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 2013;200:950–965. doi: 10.1111/nph.12423. [DOI] [PubMed] [Google Scholar]
- Chao D-Y, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis . Science. 2013;341:658–659. doi: 10.1126/science.1240561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chang SX, Anyia AO. Gene discovery in cereals through quantitative trait loci and expression analysis in water-use efficiency measured by carbon isotope discrimination. Plant Cell Environ. 2011;34:2009–2023. doi: 10.1111/j.1365-3040.2011.02397.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Marroig G. Comparing covariance matrices: Random skewers method compared to the common principal components model. Genet. Mol. Biol. 2007;30:461–469. [Google Scholar]
- Colautti RI, Barrett SCH. Population divergence along lines of genetic variance and covariance in the invasive plant Lythrum salicaria in eastern North America. Evolution. 2011;65:2514–2529. doi: 10.1111/j.1558-5646.2011.01313.x. [DOI] [PubMed] [Google Scholar]
- Culley TM, Dunbar-Wallis AK, Sakai AK, Weller SG, Mishio M, Campbell DR, Herzenach M. Genetic variation of ecophysiological traits in two gynodioecious species of Schiedea (Caryophyllaceae) New Phytol. 2006;169:589–601. doi: 10.1111/j.1469-8137.2005.01588.x. [DOI] [PubMed] [Google Scholar]
- Donovan LA, Dudley S, Rosenthal DM, Ludwig F. Phenotypic selection on leaf water use efficiency and related ecophysiological traits for natural populations of desert sunflowers. Oecologia. 2007;152:13–25. doi: 10.1007/s00442-006-0627-5. [DOI] [PubMed] [Google Scholar]
- Donovan LA, Ludwig F, Rosenthal DM, Rieseberg LH, Dudley S. Phenotypic selection on leaf ecophysiological traits in Helianthus . New Phytol. 2009;183:868–879. doi: 10.1111/j.1469-8137.2009.02916.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary Genetics of Genome Merger and Doubling in Plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Earley EJ, Ingland B, Winkler J, Tonsor SJ. Inflorescences contribute more than rosettes to lifetime carbon gain in Arabidopsis thaliana (Brassicaceae) Am. J. Bot. 2009;96:786–792. doi: 10.3732/ajb.0800149. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009;183:325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrecht BMJ, Comita LS, Condit R, Kursar TA, Tyree MT, Turner BL, Hubbell SP. Drought sensitivity shapes species distribution patterns in tropical forests. Nature. 2007;447:80–83. doi: 10.1038/nature05747. [DOI] [PubMed] [Google Scholar]
- Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- Geber M, Dawson TE. Genetic variation in stomatal and biochemical limitations to photosynthesis in the annual plant, Polygonum arenastrum . Oecologia. 1997;109:535–546. doi: 10.1007/s004420050114. [DOI] [PubMed] [Google Scholar]
- Geber M, Griffen LR. Inheritance and natural selection on functional traits. Int. J. Plant Sci. 2003;164:S21–S42. [Google Scholar]
- Hahn MA, Kleunen M, Müller-Schärer H. Increased phenotypic plasticity to climate may have boosted the invasion success of polyploid Centaurea stoebe . PLoS One. 2012;7:e50284. doi: 10.1371/journal.pone.0050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G-Y, Lucero ME, Sanderson SC, Zacharias EH, Holbrook NM. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae) New Phytol. 2013;197:970–978. doi: 10.1111/nph.12051. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Draper J, Jenkins G. Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv. Chromosome Res. 2004;12:397–403. doi: 10.1023/B:CHRO.0000034130.35983.99. [DOI] [PubMed] [Google Scholar]
- Heschel MS, Donohue K, Hausmann N, Schmitt J. Population differences and natural selection for water-use efficiency in Impatiens capensis (Balsaminaceae) Int. J. Plant Sci. 2002;163:907–912. [Google Scholar]
- Heschel MS, Sultan SE, Glover S, Sloan D. Population differentiation and plastic responses to drought stress in the generalist annual Polygonum persicaria . Int. J. Plant Sci. 2004;165:817–824. [Google Scholar]
- Heschel MS, Riginos C. Mechanisms of selection for drought stress tolerance and avoidance in Impatiens capensis (Balsaminaceae) Am. J. Bot. 2005;92:37–44. doi: 10.3732/ajb.92.1.37. [DOI] [PubMed] [Google Scholar]
- IBI. Update on genomics and basic biology of Brachypodium . Trends Plant Sci. 2014;19:414–418. doi: 10.1016/j.tplants.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Ivey C, Carr D. Tests for the joint evolution of mating system and drought escape in Mimulus . Ann. Bot-London. 2012;109:583–598. doi: 10.1093/aob/mcr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Li W-L, Berlyn GP, Ashton PMS. Polyploids and their structural and physiological characteristics relative to water deficit in Betula papyrifera (Betulaceae) Am. J. Bot. 1996;83:15–20. [Google Scholar]
- López-Alvarez D, López-Herranz ML, Betekhtin A, Catalán P. A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae) PLoS One. 2012;7:e51058. doi: 10.1371/journal.pone.0051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, Liu J, Yub X, Jiang Y. Natural variation of drought response in Brachypodium distachyon . Physiol. Plantarum. 2011;141:19–29. doi: 10.1111/j.1399-3054.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. Genome duplication and the evolution of physiological responses to water stress. New Phytol. 2009;184:721–731. doi: 10.1111/j.1469-8137.2009.02997.x. [DOI] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae) New Phytol. 2012;193:797–805. doi: 10.1111/j.1469-8137.2011.03988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron JL, Elmendorf SC, Vilà M. Contrasting plant physiological adaptation to climate in the native and introduced range of Hypericum perforatum . Evolution. 2007;61:1912–1924. doi: 10.1111/j.1558-5646.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- Martin SL, Husband BC. Whole genome duplication affects evolvability of flowering time in an autotetraploid plant. PLoS One. 2012;7:e44784. doi: 10.1371/journal.pone.0044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Husband BC. Adaptation of diploid and tetraploid Chamerion angustifolium to elevation but not local environment. Evolution. 2013;67:1780–1791. doi: 10.1111/evo.12065. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleitropy contributes to genetic correlations among ecological traits. Mol. Ecol. 2003;12:1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Davidson A. Adaptive phenotypic plasticity and plant water use. Functional Plant Biology. 2010;37:117–127. [Google Scholar]
- Nuismer SL, Cunningham BM. Selection for phenotypic divergence between diploid and autotetraploid Heuchera grossulariifolia . Evolution. 2005;59:1928–1935. [PubMed] [Google Scholar]
- Oswald BP, Nuismer SL. Neopolyploidy and diversification in Heuchera grossulariifolia . Evolution. 2010;65:1667–1679. doi: 10.1111/j.1558-5646.2010.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. The Evolutionary Consequences of Polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Paccard A, Fruleux A, Willi Y. Latitudinal trait variation and responses to drought in Arabidopsis lyrata . Oecologia. 2014;175:577–587. doi: 10.1007/s00442-014-2932-8. [DOI] [PubMed] [Google Scholar]
- Pacheco-Villalobos D, Hardtke CS. Natural genetic variation of root system architecture from Arabidopsis to Brachypodium: towards adaptive value. Philos. T. R. Soc. B. 2012;367:1552–1558. doi: 10.1098/rstb.2011.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C. Polyploids integrate genomic changes and ecological shifts. New Phytol. 2012;193:297–300. doi: 10.1111/j.1469-8137.2011.04008.x. [DOI] [PubMed] [Google Scholar]
- Prentice IC, Meng T, Wang H, Harrison SP, Ni J, Wang G. Evidence of a universal scaling relationship for leaf CO2 drawdown along an aridity gradient. New Phytol. 2011;190:169–180. doi: 10.1111/j.1469-8137.2010.03579.x. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Polyploidy and ecological adaptation in wild yarrow. P. Natl. Acad. Sci. USA. 2011;108:7096–7101. doi: 10.1073/pnas.1016631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson IH. Chromosome numbers in Brachypodium Beauv. (Gramineae) Genetica. 1981;56:55–60. [Google Scholar]
- Sgrò CM, Hoffmann AA. Genetic correlations, tradeoffs and environmental variation. Heredity. 2004;93:241–248. doi: 10.1038/sj.hdy.6800532. [DOI] [PubMed] [Google Scholar]
- Sherrard ME, Maherali H. The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution. 2006;60:2478–2489. [PubMed] [Google Scholar]
- Sherrard ME, Maherali H, Latta RG. Water stress alters the genetic architecture of functional traits associated with drought adaptation in Avena barbata . Evolution. 2009;63:702–715. doi: 10.1111/j.1558-5646.2008.00580.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since Plant speciation . New Phytol. 2004;161:173–191. [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS. What we still don’t know about polyploidy. Taxon. 2010;59:1387–1403. [Google Scholar]
- Schranz ME, Osborn TC. De novo variation in life-history traits and responses to growth conditions of resynthesized polyploid Brassica napus (Brassicaceae) Am. J. Bot. 2004;91:174–183. doi: 10.3732/ajb.91.2.174. [DOI] [PubMed] [Google Scholar]
- Schwartz CJ, Doyle MR, Manzaneda AJ, Rey PJ, Mitchell-Olds T, Amasino RM. Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon . Bioenerg. Res. 2010;3:38–46. [Google Scholar]
- Tardieu F, Tuberosa R. Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 2010;13:206–212. doi: 10.1016/j.pbi.2009.12.012. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubešová M, Pyšek P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot-London. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier UA, Broennimann O, Normand S, Guisan A, Schaffner U, Steinger T, Müller-Schärer H. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa Ecology. 2009;90:1366–1377. doi: 10.1890/08-0420.1. [DOI] [PubMed] [Google Scholar]
- Tuberosa R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012;3:e347. doi: 10.3389/fphys.2012.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Tuna M, Budak H, Huo N, Gu YQ, Steinwand MA. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon . BCM Plant Biol. 2009;9:88. doi: 10.1186/1471-2229-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DA, Edwards GE. Effects of polyploidy on photosynthesis. Photosynthesis Research. 1993;35:135–147. doi: 10.1007/BF00014744. [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. P. Natl. Acad. Sci. USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Nutter LI, Willis JH. Natural variation for drought-response traits in the Mimulus guttatus species complex. Oecologia. 2010;162:23–33. doi: 10.1007/s00442-009-1448-0. [DOI] [PubMed] [Google Scholar]
- Xu Y, This D, Pausch RC, Vonhof WM, Coburn JR, Comstock JP, McCouch SR. Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theor. Appl. Genet. 2009;118:1065–1081. doi: 10.1007/s00122-009-0963-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.